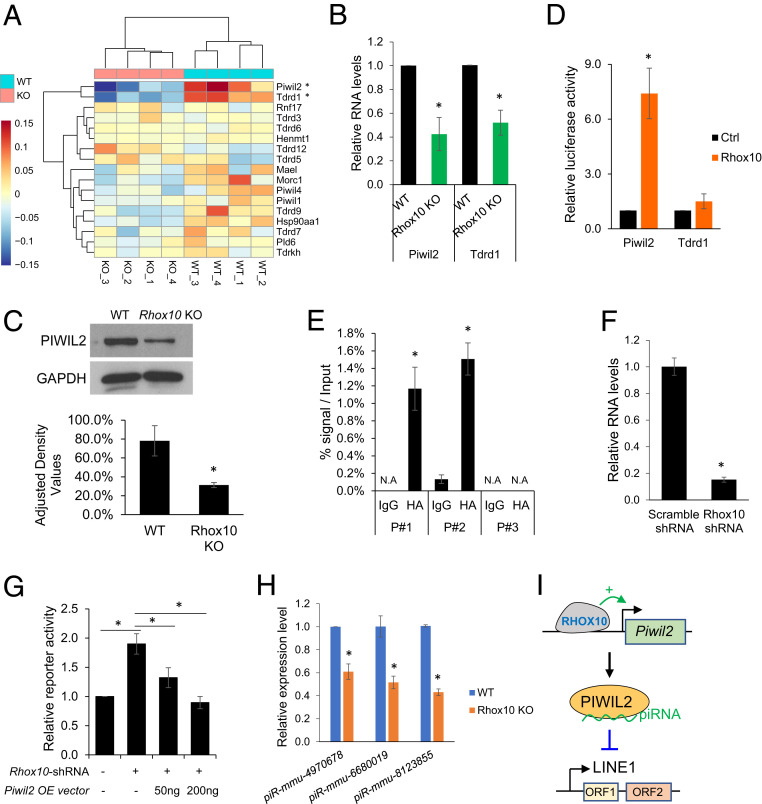
Fig. 3.
RHOX10 suppresses LINE1 expression via regulating Piwil2 expression. (A) Heatmap of piRNA pathway gene expression determined by the RNA-seq analysis shown in Fig. 2. *, q < 0.05, Basemean > 10, |Log2FC| > 0.25. (B) qPCR validation of Rhox10-mediated regulation of Piwil2 and Tdrd1 expression in germ cells from E16.5 Rhox10-null; Oct4-eGFP+/+ (KO) and Oct4-eGFP+/+ (WT) mice (n = 3). *P < 0.05. (C, Top) Western blot analysis of PIWIL2 protein expression in testes lysates from E16.5Rhox10-null (KO) and littermate WT mice. (Bottom) PIWIL2 quantification when normalized to GAPDH (n= 3). *P< 0.05.(D) Luciferase analysis of Piwil2 or Tdrd1 promoter activity in HEK-293T cells cotransfected with a luciferase reporter vector harboring the Piwil2 or Tdrd1 promoter and a Rhox10-expression vector or an empty expression vector as a control (Ctrl) (n = 4). *P < 0.05. (E) ChIP-qPCR analysis of GS cells transfected with an expression vector encoding HA-tagged RHOX10 and analyzed using an HA antibody. Endogenous RHOX10 was knocked down using a Rhox10-shRNA lentivirus (>80% efficiency) (51). IgG antibody was used as a negative control. The values shown are the DNA signal in the immunoprecipitation samples relative to the input. (n = 3). *P < 0.05. (F) qPCR analysis of Rhox10 expression in GS cells transduced with a Rhox10-shRNA lentivirus or a scramble-shRNA lentivirus as a control (n = 2). *P < 0.05. (G) Luciferase analysis of LINE1 transposition activity in GS cells cotransfected with the ORFeus mouse LINE1 reporter, a Piwil2 expression vector, and a Rhox10-shRNA vector, as shown (n = 3). *P < 0.05. (H) TaqMan‐qPCR analysis of the indicated piRNAs in FACS-purified germ cells from E16.5 Rhox10-null; Oct4-eGFP+/+ (Rhox10 KO) and littermate Oct4-eGFP+/+ (WT) mice (n = 3). *P < 0.05. U6 small nuclear RNA levels were used for normalization. (I) Model: the RHOX10 transcription factor activates Piwil2 transcription to suppresses LINE1 elements.