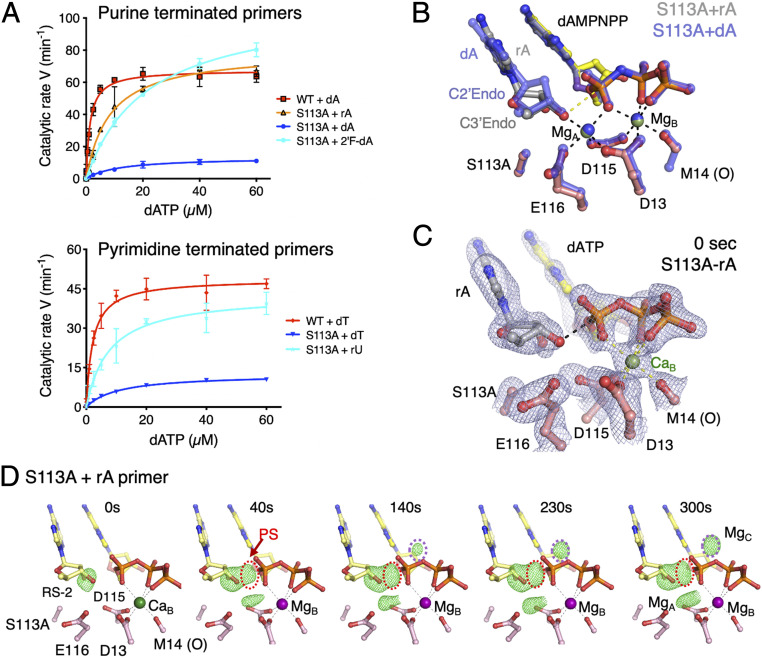
Fig. 4.
RNA-terminated primer rescues the defects of S113A Pol η. (A) Steady-state kinetic profiles of WT and S113A mutant Pol η extending primers terminating in dA, rA, or 2′-fluoro-dA (Top) and dT or rU (Bottom). Kinetic parameters for each profile are listed in Table 1. Error bars represent standard deviation of triplicate measurements. (B) Superimposed structures of S113A Pol η bound to the dA- or rA-primer and dAMPNPP. The dA-primer structure is shown in semitransparent blue sticks and the rA-primer structure is shown in colored sticks (yellow dAMPNPP, salmon protein residues, and gray primer). The sugar pucker of the terminal primer base is labeled. (C) The 0-s structure of S113A Pol η bound to the rA-primer with the 2Fo−Fc map (1σ) superimposed. The rA adopts the C3′-endo conformation and is reaction-ready (RS-2) prior to Mg2+ binding in the A site. (D) The reaction time course of S113A extending the rA-primer. Reaction time course of the rA-primer extension by S113A. Fo(S113A-rA, 0–300 s)−Fc(WT, GS) maps (3.5σ) with C3′ and O3′ omitted are superimposed onto the 0-s S113A Pol η–rA-primer complex at 0 s. The electron densities for the product and Mg2+C are circled in red and purple, respectively.