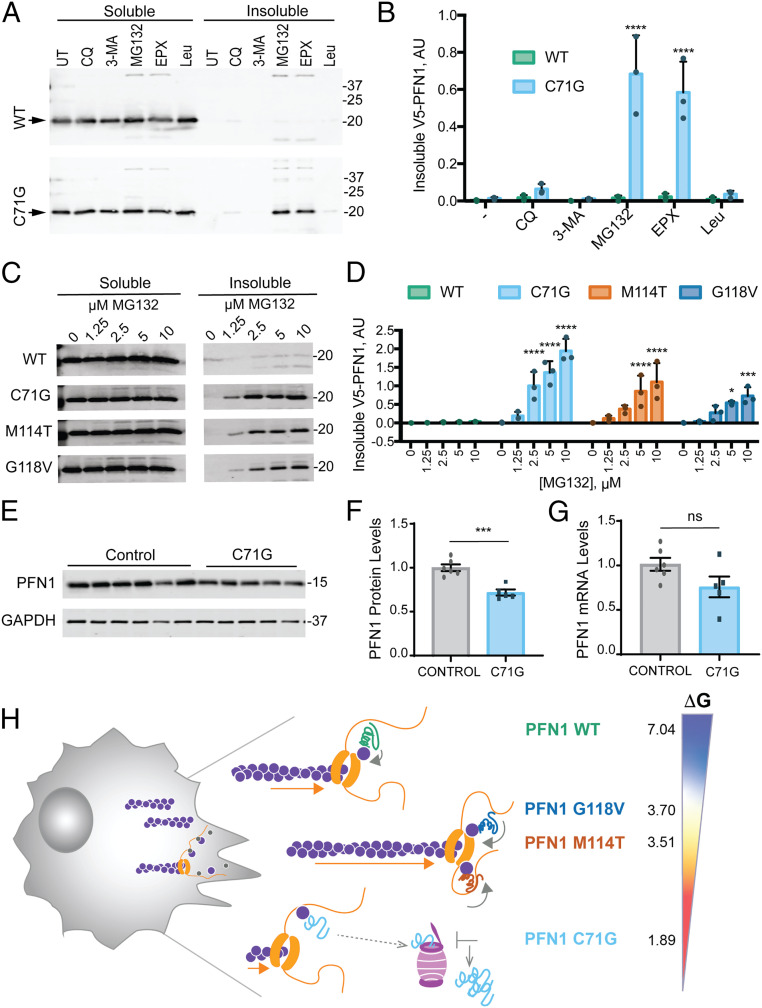
Fig. 4.
The PFN1 C71G variant is robustly targeted by the proteasome and exhibits reduced steady-state protein expression levels. (A) Western blot analysis of detergent soluble and insoluble fractions from HeLa cells expressing V5-PFN1 WT or C71G treated with proteasome or autophagy/lysosome inhibitors for 24 h prior to cell lysis. Soluble and insoluble fractions were processed on the same gel. Immunoblots were probed with anti-V5 antibody; arrowheads indicate V5-PFN1 WT and C71G. Abbreviations: UT: untreated, CQ: chloroquine (100 μM), 3-MA: 3-methyladenine (5 mM), MG: MG132 (10 μM), EPX: epoxomicin (10 μM), and Leu: leupeptin (100 μM). (B) Quantification of insoluble fractions from A (n = 3 biological replicates, mean ± SD, two-way ANOVA with Dunnett’s multiple comparison test for difference from untreated, ****P < 0.0001). (C) The indicated V5-PFN1 cell lines were treated with increasing concentrations MG132 for 24 h and processed as described in A. (D) Quantification of insoluble fractions from C (n = 3 biological replicates, mean ± SD, two-way ANOVA with Dunnett’s multiple comparison test for difference from WT, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (E) Western blot analysis of endogenous, untagged PFN1 protein levels and GAPDH loading control from immortalized human lymphoblast cells derived from controls and ALS patients harboring the C71G PFN1 mutation. (F) Quantification of E. For each biological replicate, PFN1 protein levels were normalized to the average level of the control cell lines. Each data point represents the average protein levels of n = 2 biological replicates for individual cell lines. Bar graphs show mean ± SEM among lymphoblast lines. Statistics were determine using student’s t test (***P < 0.001). (G) Relative mRNA levels of PFN1 normalized to the average level of the control cell lines. Each data point represents the average mRNA levels of n = 2 to 3 biological replicates for individual lines. Bar graphs show mean ± SEM among lymphoblast lines. (H) Proposed model for the effects of ALS-linked mutations on PFN1 in the context of formin-induced actin polymerization. Our MD simulations and the lower free energy of folding [ΔG°; determined by Boopathy et al. (9)] indicate regions within PFN1 that contact both actin and poly-Pro are more flexible in ALS-linked variants compared with PFN1 WT. This conformational flexibility may contribute to enhanced formin binding and processivity (depicted by orange arrows), potentially by affecting the ring complex (i.e., PFN1-actin bound to FH1 in contact with the FH2-bound barbed end). PFN1 C71G appears to be the least competent at promoting formin-induced actin polymerization. This variant also exhibits the lowest ΔG° (kcal/mol); 4% of PFN1 C71G exist in the unfolded state, compared with <0.3% for M114T and G118V and <0.0007% for WT. These data are consistent with the C71G variant adopting an unfolded and/or misfolded conformation that is more robustly targeted to the proteasome. Compared with PFN1 WT, all ALS-linked variants dysregulate formin-induced actin polymerization.