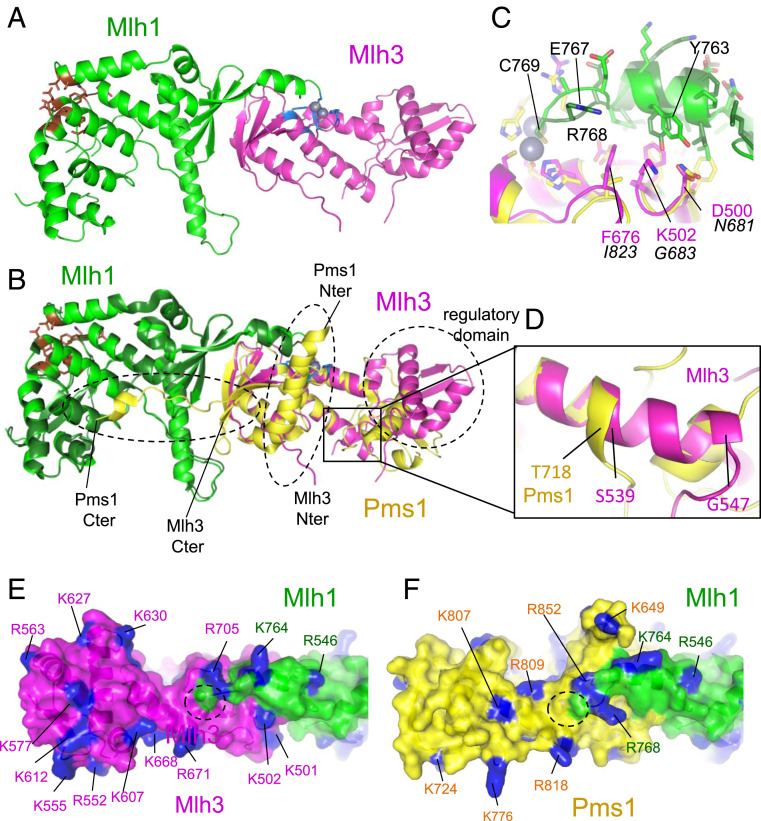
Fig. 1.
Crystal structure of MutLγ(CTD). (A) Overall view of the MutLγ(CTD) heterodimer (Mlh1 and Mlh3 are colored respectively in green and magenta). The endonuclease site is colored in blue with the two zinc atoms in sphere representation and the Exo1 binding site in brown. (B) Superimposition of the MutLα(CTD) on the MutLγ(CTD). Mlh1 and Pms1 in MutLα are colored respectively in dark green and yellow. The three main regions of Pms1(CTD) that differ from Mlh3(CTD) are highlighted with dashed lines: 1) the regulatory domains, 2) the first residues of Pms1 and Mlh3 CTDs (the position of the first residues of the two CTDs are indicated [“Nter”]), and 3) the last residues of both CTDs (“Cter”). (C) The last residue Cys769 of Mlh1 adopts the same position in the endonuclease site in both complexes. The preceding residues of Mlh1 adopt a slightly different conformation when they are in contact with Mlh3 residues (numbers in magenta) or Pms1 ones (numbers in italic). (D) The helix αA of Mlh3 located in the hinge between the regulatory and dimerization domains is two turns longer in Mlh3 than in Pms1 pushing away the regulatory domain of Mlh3 compared to Pms1. The region zoomed corresponds to the black rectangle shown in B. The distribution of the basic residues (colored in blue) around the Mlh3 (E) and Pms1 (F) active sites is different (active site located with dashed lines). The position of the regulatory domain in Mlh3 also differs from the one observed in Pms1 resulting in different shapes of the surface surrounding the endonuclease sites of Mlh1-Mlh3 and Mlh1-Pms1.