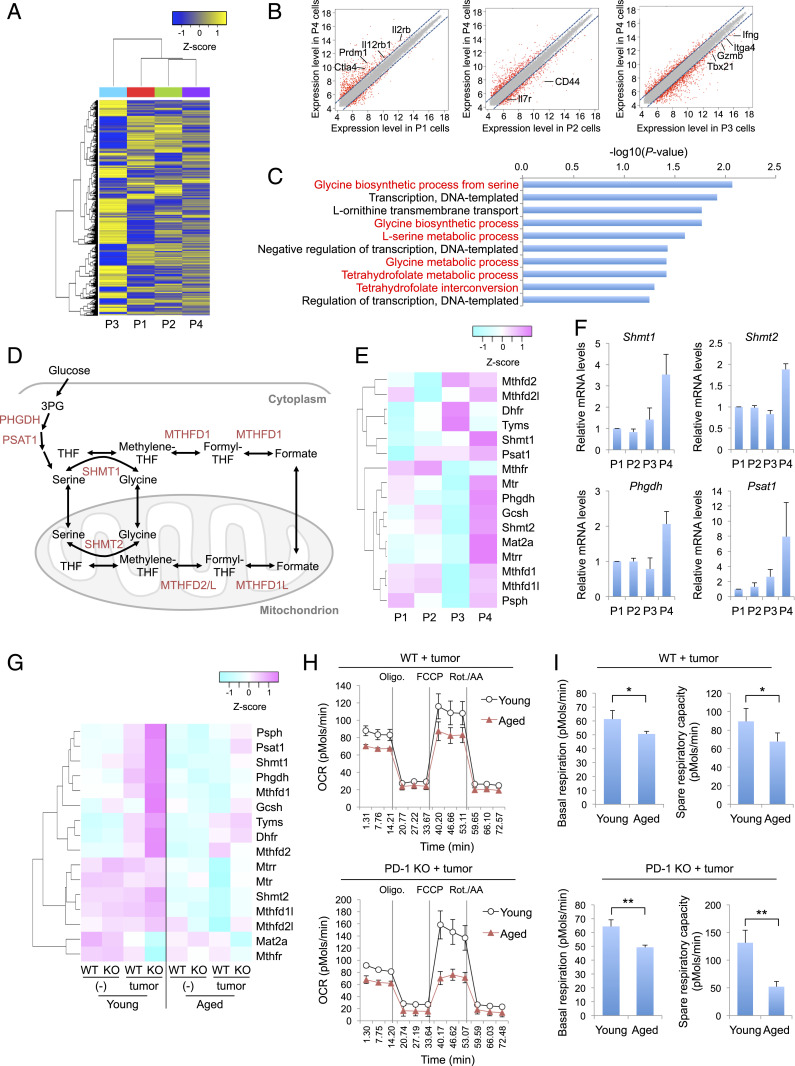
Fig. 3.
Increased expression of 1C-metabolism–related genes in P4 cells. (A–E) Microarray analysis of P1, P2, P3, and P4 cells sorted from young PD-1 KO mice (1–3 mo old; nine mice pooled). (A) Hierarchical clustering heat map of all detected genes. (B) Scatter plots represent normalized log intensities of individual probes. The dashed lines indicate log 2-fold change. Genes previously linked to CD8+ T cell activation and differentiation are listed. (C) Top 10 enriched gene ontology (GO) terms from the up-regulated genes in P4 cells. GO terms related to 1C metabolism are highlighted in red. (D) Schematic of 1C-metabolic pathways. THF, tetrahydrofolate. (E) Heat map showing the expression of 1C-metabolism–related genes in CD8+ T cell subsets. (F) Relative mRNA expression of 1C-metabolism–related genes in CD8+ T cell subsets from splenocytes of young PD-1 KO mice. (G) Microarray analysis of WT or PD-1 KO CD8+ T cells in PLNs or DLNs of young (2 mo old, three mice pooled) or aged (17 mo old, six mice pooled) mice with or without MC38 tumors (day 9). Heat map showing the expression of 1C-metabolism–related genes in the indicated cells. (H and I) Oxygen consumption rate (OCR) of WT or PD-1 KO CD8+ T cells from DLNs of young (2–3 mo old) or aged (15–19 mo old) mice was measured using a Seahorse XFe96 analyzer. OCR trace (H) and basal respiration and spare respiratory capacity were calculated from OCR values (I). *P < 0.05; **P < 0.01 (two-tailed unpaired Student’s t test). Data are presented as the mean ± SEM (n = 4).