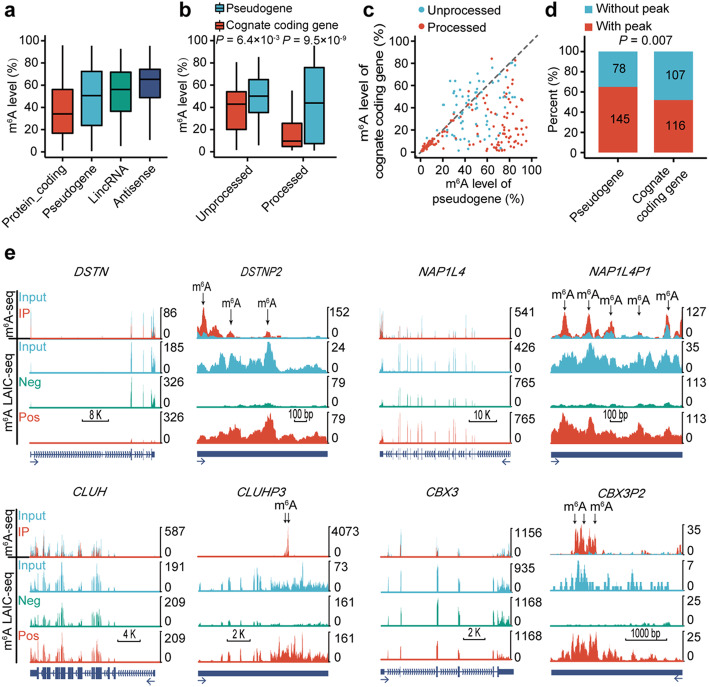
Fig. 1.
Comparisons of m6A between pseudogenes and their cognate protein-coding genes in GM12878 cell line. a Box plot comparing the m6A levels among different biotypes. b Box plot comparing the m6A levels between pseudogenes and their cognate protein-coding genes for processed pseudogenes and unprocessed pseudogenes respectively. c Scatter plot comparing the m6A levels between pseudogenes and their cognate protein-coding genes for processed pseudogenes and unprocessed pseudogenes respectively. d Stacked bar plot comparing the percent of pseudogenes and their cognate protein-coding genes with or without m6A peaks. The frequencies were displayed in the corresponding boxes. e UCSC genome browser tracks of m6A-seq and m6A-LAIC-seq data showing representative examples of pseudogenes and their cognate protein-coding genes. Read-coverage tracks of input, m6A-negative, and m6A-positive fractions of m6A-LAIC-seq shown along with overlay tracks of m6A-seq (red for RIP and cyan for input; predicted m6A sites in m6A peaks are indicated by arrows). Read coverage (y-axis) of m6A negative and m6A positive are normalized as previously described [28] to reflects the calculated m6A levels (i.e., equal signals in m6A positive (eluate) versus m6A negative (supernatant) indicate m6A levels of 50%), while input and m6A-seq tracks are shown for optimal viewing