Figure 6. RIME analysis of GATA6 interacting proteins.
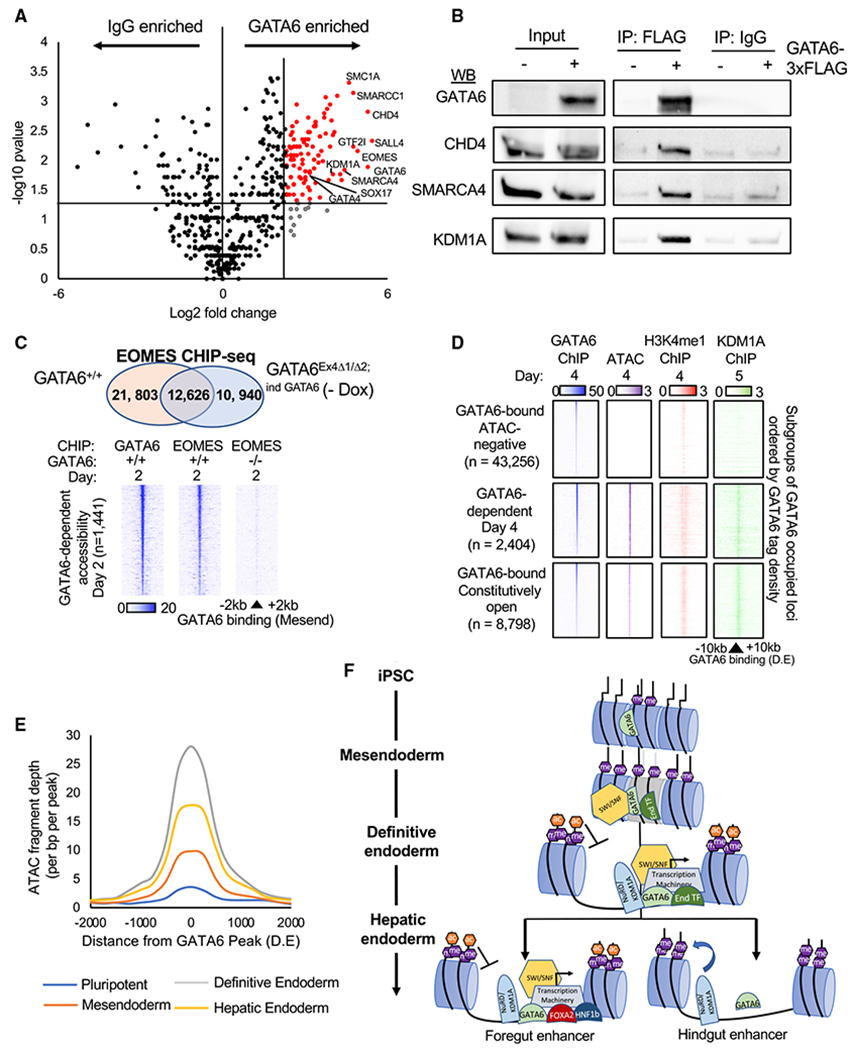
(A) Volcano plot of RIME-detected proteins with more than one spectral count and more than one unique peptide fragment. GATA6 interactome: 5-fold enriched over IgG controls, p < 0.05, n = 2. See Table S5.
(B) Validation of RIME proteins by western blot analysis of GATA6, CHD4, SMARCA4, and KDM1A following immunoprecipitation of GATA6-3× FLAG protein in GATA6Ex4Δ1/Δ2;ind GATA6 mesendoderm cultures differentiated with and without doxycycline.
(C) Venn diagram depicting overlap of peaks from EOMES ChIP-seq completed in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind GATA6 cells without doxycycline differentiated to mesendoderm (upper panel). Heatmaps of EOMES and GATA6 ChIP-seq intensity in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind GATA6 cells without doxycycline at regions of rapid GATA6-dependent chromatin remodeling (day 2) (lower panel).
(D) Published KDM1A ChIP-seq (Vinckier et al., 2020) from similar differentiation stage (day 5; foregut) overlapped with definitive endoderm GATA6 binding sites. GATA6 (blue), KDM1A (green), H3K4me1 (red), and ChIP-seq and ATAC-seq (purple) intensity displayed at subgroups of GATA6-bound sites with different endoderm accessibility profiles.
(E) Temporal profile of ATAC-seq data fragment depth during the hepatic specification at sites of GATA6-dependent chromatin accessibility. See Table S6.
(F) Proposed model of GATA6-dependent chromatin modulation during endoderm formation and specification. End transcription factors (TFs): EOMES, SOX17, and FOXA2.
