Extended Data Fig. 7. Species-wide SNP-based relatedness of divergent regions is in agreement with long-read sequencing results.
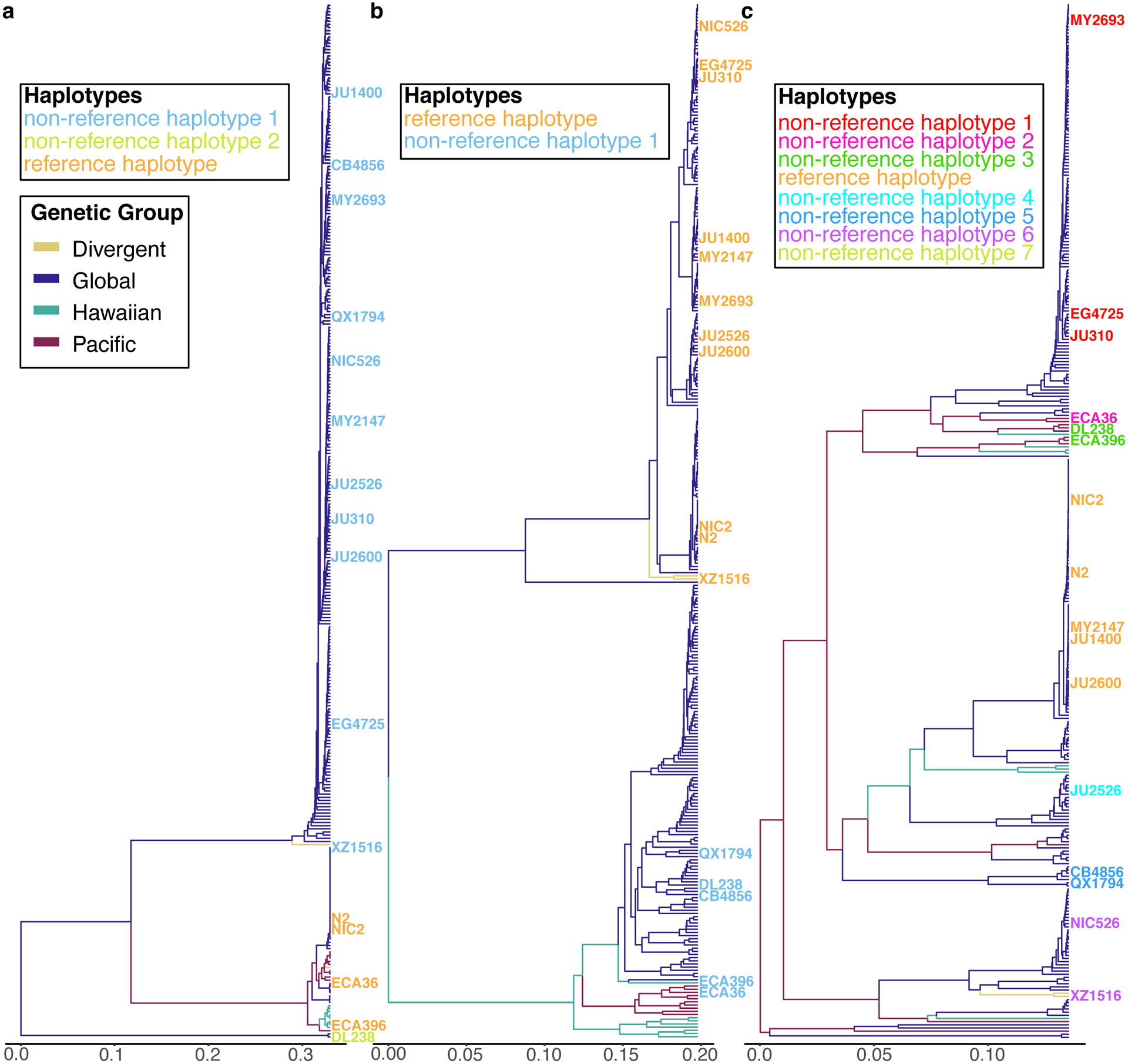
The inferred for the C. elegans species-wide relatedness for the hyper-divergent regions that span (a) II:3,667,179–3,701,405, (b) I:2,318,291–2,381,851, and (c) V:20,193,463–20,267,244 are shown. The x-axis represents the dissimilarity of the fraction of identity-by-state in the region. For a-c, the isotype names are colored to match the haplotypes defined by long-read sequence data in Fig. 5 and Extended Fig. 8,9, respectively. The branch colors correspond to the species-wide genetic groups identified by PCA analysis in Fig. 1c.
