Fig. 2 |. Characterization of hyper-divergent regions at the isotype level.
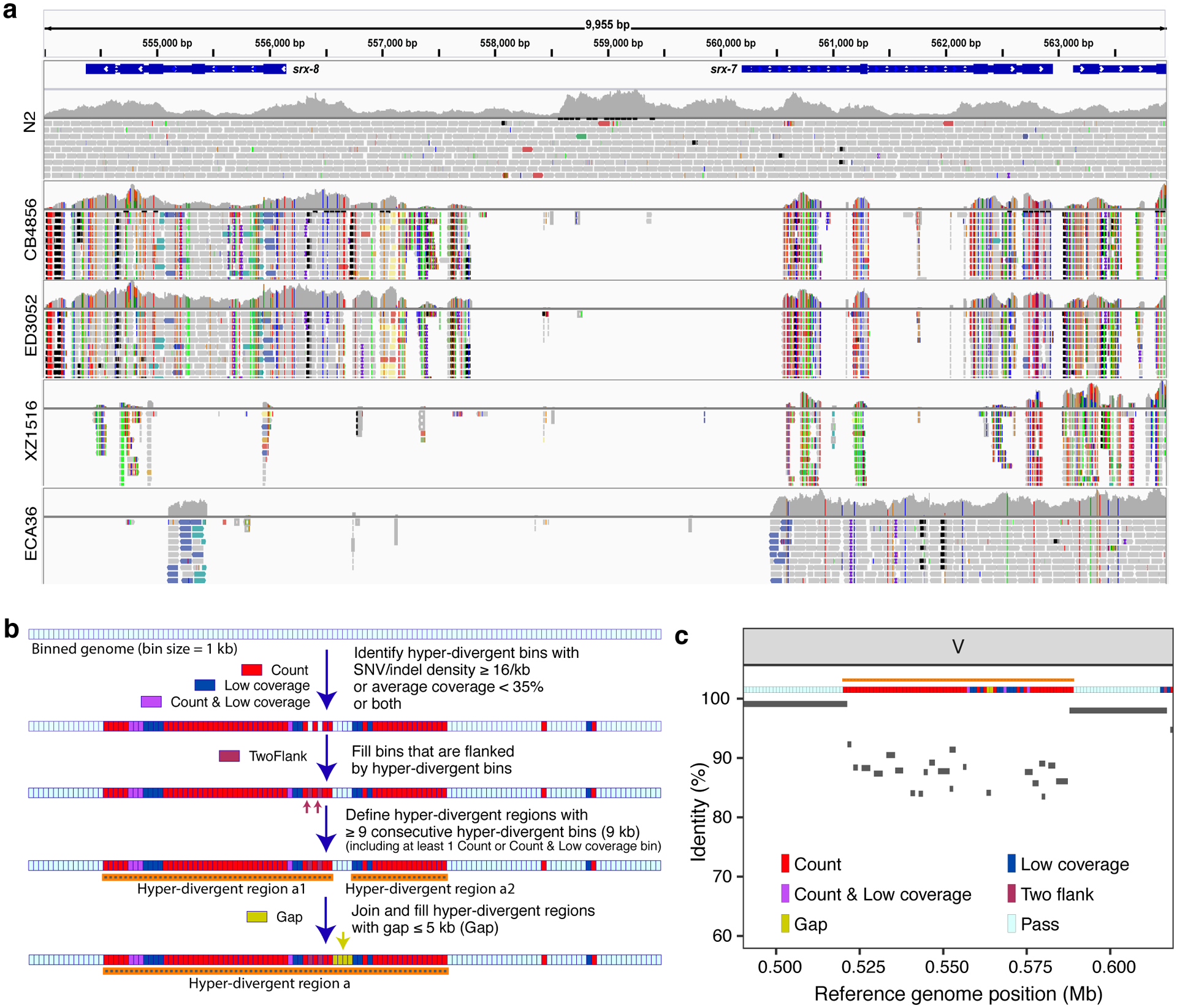
(a) Short-read alignments from five isotypes (N2, CB4856, ED3052, XZ1516, and ECA36) to the N2 reference genome (WS245) for a region of chromosome V (V:554,000–564,000) are shown. Genes (srx-7 and srx-8) in the interval are shown at the top. For each isotype, the top panel shows the coverage of genomic positions and the bottom panel shows aligned short-reads at genomic positions (gray: normal reads, red: reads with putative deletion, blue: reads with putative insertion, navy and turquoise: reads with putative inversion, green: reads with putative duplication or translocation). Colored vertical lines indicate mismatched bases at the position.
(b) The workflow for the characterization of hyper-divergent regions at the isotype level is shown (See Methods).
(c) An example plot showing the short-read based characterization of a hyper-divergent region and the percent sequence identity from a long-read alignment. The orange horizontal line shows a hyper-divergent region (V:520,000–589,000) in the locus (V:490,000–619,000) of CB4856 classified using this approach. The multi-colored horizontal rectangle shows classifications of genomic bins in the locus. Gray bars correspond to the alignments from CB4856 long-read sequences to the N2 reference genome (WS245), and identities of alignments are shown on the y-axis.
