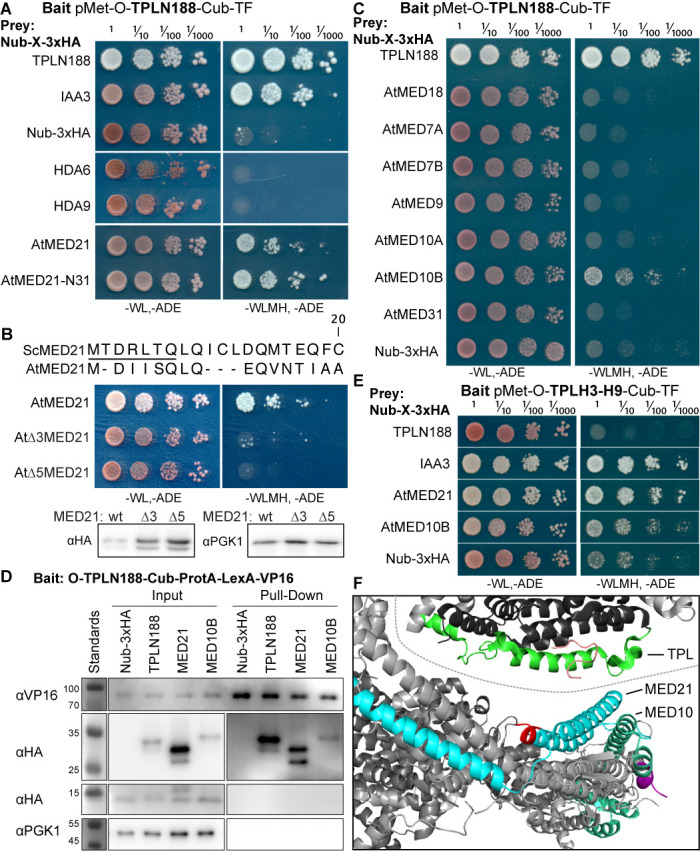
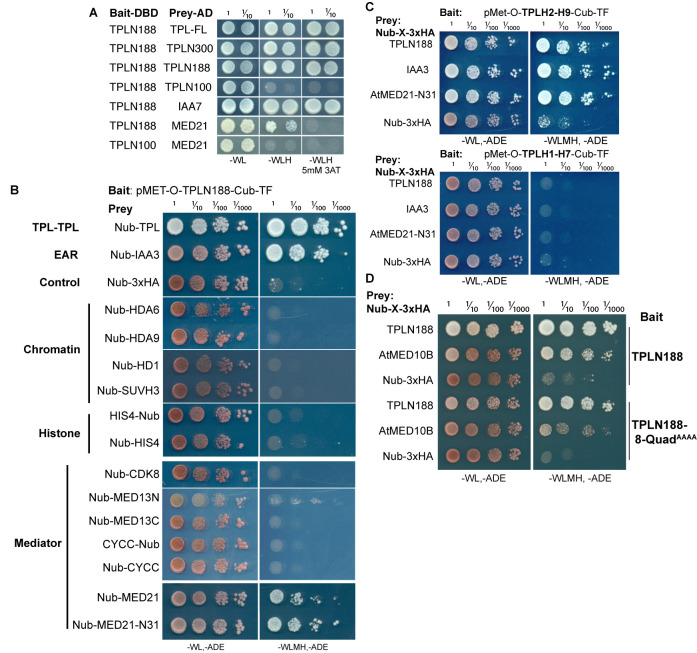
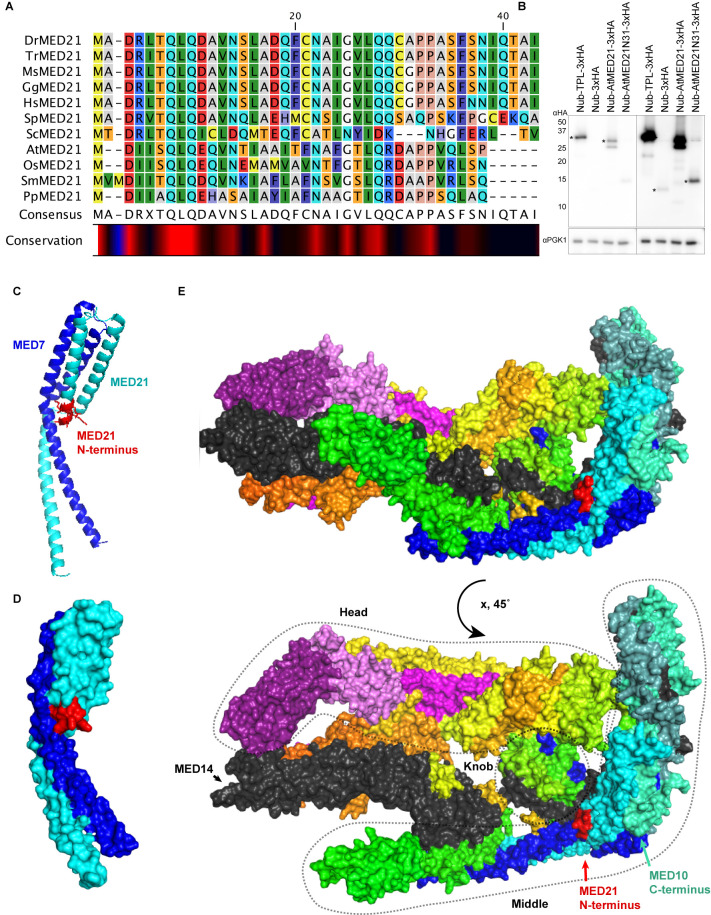
Figure 2. The Helix 8 repression domain of TPL directly interacts with AtMED21 and AtMED10B.
(A–C, E) Cytoplasmic split-ubiquitin system (CytoSUS) assays with candidate interacting proteins. Nub-3xHA is the N-terminal fragment of ubiquitin expressed with no fusion protein and is used as a negative control. Each prey protein is from Arabidopsis. -WL, -ADE: dropout lacking Trp, Leu, and Ade (growth control); -WLMH, -ADE: dropout lacking Trp, Leu, His, Met, and Ade (selective media). The plating for each panel was performed at the same day, white lines are provided when plates were cropped for clarity. (B) Alignments of the Arabidopsis (At) and Saccharomyces (Sc) MED21 proteins are shown above cytoSUS assays with the same bait shown in (A). Western blots below the colonies indicated that AtMED21 N-terminal Δ3 and Δ5 are well expressed in assay conditions. (C) CytoSUS assays with selected Mediator proteins in the middle module. (D) The TPL-ProteinA-TF fusion protein can pull down TPL, AtMED21, and AtMED10B from yeast extracts using IgG-beads. Detection of the VP16 transcriptional activator demonstrates enrichment of the fusion protein (αVP16). Each prey protein is detected via the 3xHA tag (αHA), and efficacy of purification was judged by PGK1 depletion (αPGK1). (E) A TPL-N truncation lacking the LisH domain (TPLH2-H9) could still interact with the AtMED21-N31 truncation. This bait construct interacted with IAA3, but only minimally with the negative control (free Nub-3xHA). (F) Yeast Mediator (bottom, 5N9J) and AtTPL (top, 5NQV) manually juxtaposed to compare relative domain sizes and feasibility of a TPL-MED21-MED10B interaction. TPL Helix 8–9 is colored green. MED21 is colored aqua, with the N-terminus colored red, and the IAA27 EAR peptide in orange. MED10 is colored teal, with the C-terminus colored purple. The dotted line indicates the border between TPL and Mediator structures.