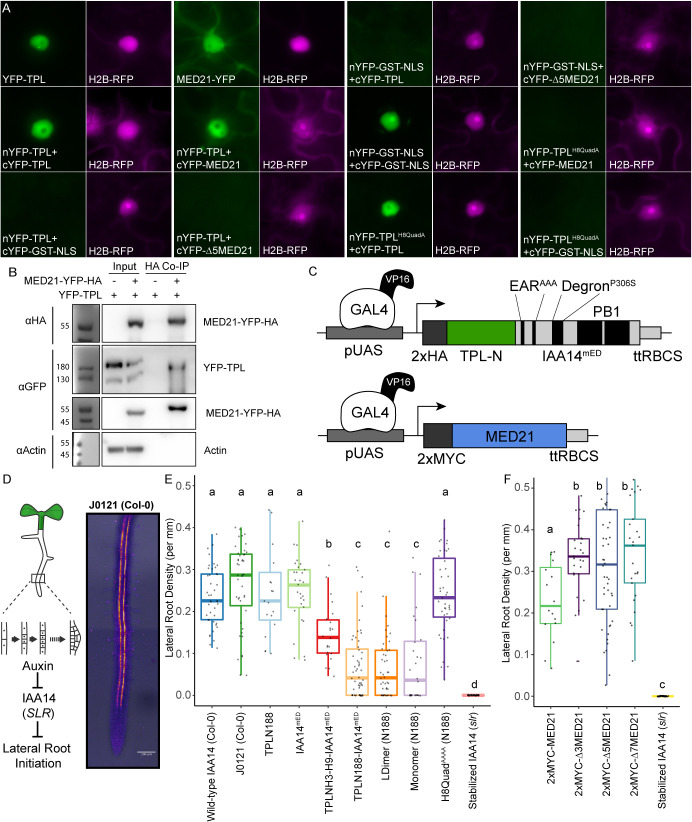
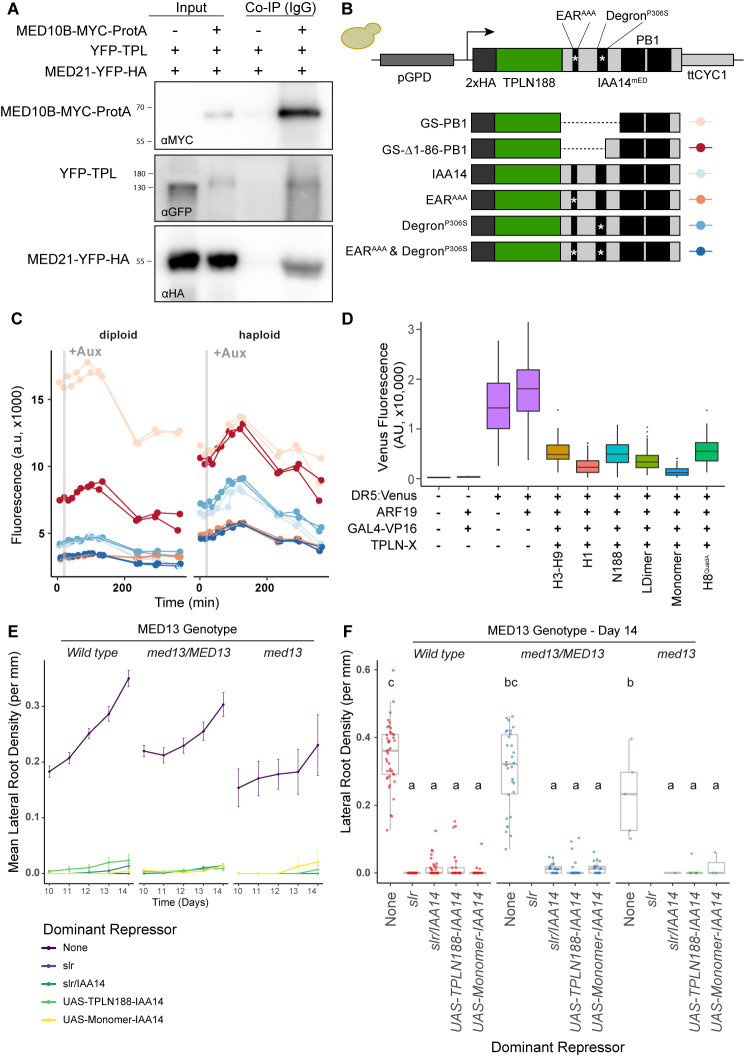
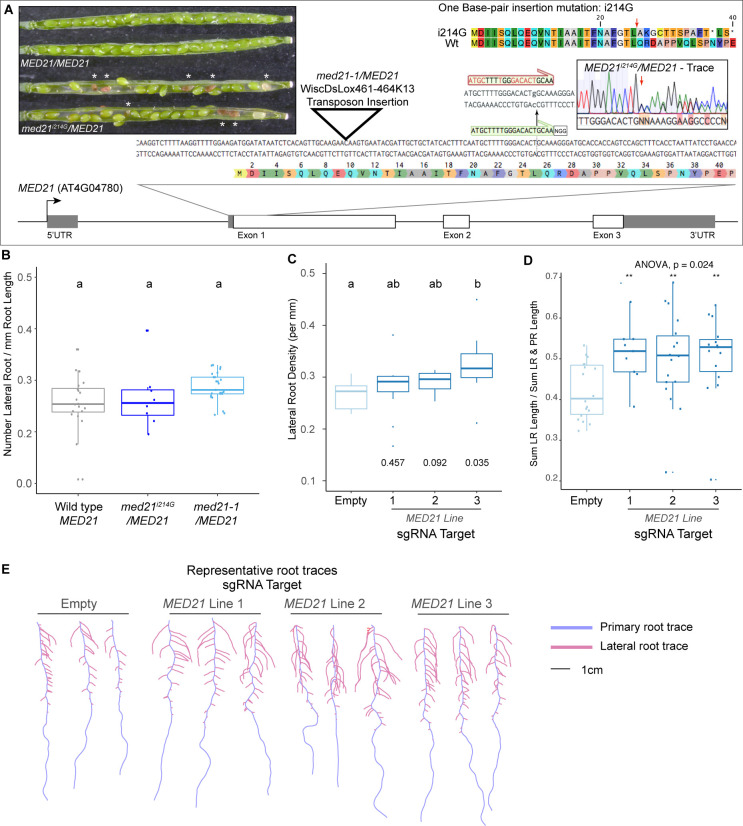
Figure 6. The TPL CRA repression domain behaves similarly in yeast and plants.
(A) Bimolecular fluorescence complementation assay performed in tobacco. Each image is an epi-fluorescent micrograph taken at identical magnification from tobacco epidermal cells at 2 days post injection. The YFP image is colored green (left panel). p35S:H2B-RFP was used as a control and is false-colored magenta (right panel). (B) Co-immunoprecipitation of MED21 and TPL from tobacco leaves. MED21-YFP-HA was immunoprecipitated using anti-HA, and YFP-TPL was detected using the YFP fusion. Actin was used to demonstrate that the purification had removed non-specific proteins. Numbers on the left of blots indicate sizes of protein standards in kilodaltons. (C) Design of UAS-TPL-IAA14mED and UAS-MED21 constructs. Mutation of the conserved lysine residues in the EAR domain disrupted potential interactions with endogenous TPL/TPR proteins. The IAA14 degron has been mutated (P306S) to render it auxin insensitive. UAS: upstream activating sequence; ttRBCS: Rubisco terminator sequence. (D) Auxin-induced degradation of IAA14 is absolutely required for initiation of lateral root development (cartoon, left). An enhancer trap line (J0121) expresses GAL4-VP16 and UAS-GFP in in xylem pole pericycle cells. (E) N-terminal domains of TPL were sufficient to repress the development of lateral roots in Arabidopsis seedlings. The density of emerged lateral roots was measured in T1 seedlings at 14 days after germination. (F) N-terminal deletions in AtMED21 were sufficient to dominantly increase the development of lateral roots in Arabidopsis seedlings. The density of emerged lateral roots was measured in T1 seedlings at 14 days after germination. (E, F) Lowercase letters indicate significant difference (ANOVA and Tukey HSD multiple comparison test; p<0.001).