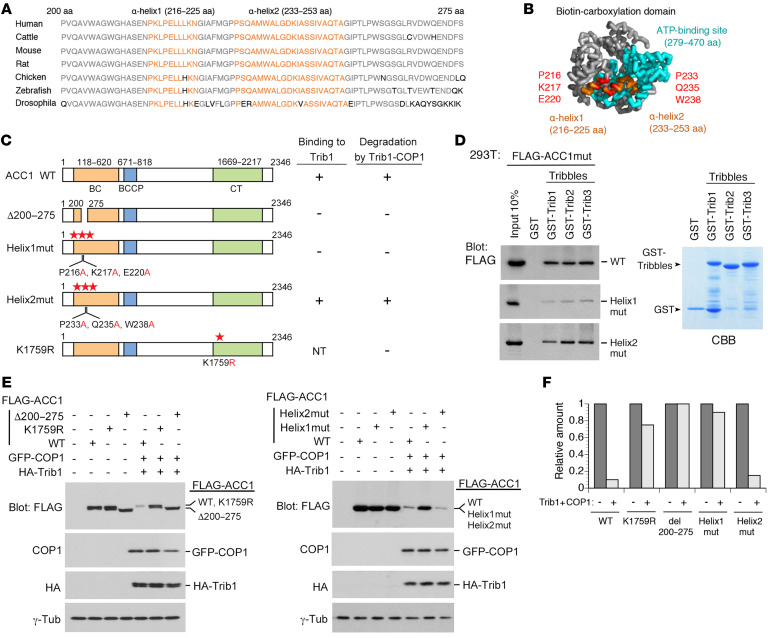
Figure 3. Construction of ACC1 point mutants resistant to Trib1-COP1–mediated degradation.
(A) Sequence alignment of residues 200–275 of human ACC1 with compatible residues of various species. The region includes two α-helices (Helix1: residues 216–225; Helix2: residues 233–253) marked in orange and other conserved residues shown in gray. (B) Structure of the biotin carboxylase (BC) domain of ACC1. Helix1 and Helix2 (in orange) are positioned at the outside of the ATP-binding site (in blue). Mutants of Helix1 and Helix2 were constructed by replacement of 3 residues positioned at the interaction surfaces with alanine (Helix1mut: P216A, K217A, and E220A; Helix2mut: P233A, Q235A, and W238A; shown in red). This model is generated by the human ACC1 full crystal structure (PDB:6G2D) with PyMOL (http://www.pymol.org). (C) Schematic representation of ACC1 point mutants for the ACC1-Trib1 binding and ubiquitination sites. The results of Trib1 binding and degradation are summarized on the right. NT, not tested. (D) GST-control and all GST-Tribbles (GST-Trib1, GST-Trib2, and GST-Trib3) fusion proteins were incubated with 293T cell lysates containing FLAG-ACC1WT, Helix1mut, and Helix2mut. Bound proteins were detected by immunoblotting with an antibody against a FLAG epitope. GST-Tribbles–fused proteins were visualized by CBB staining to evaluate their amounts. (E and F) Helix1mut and K1759R are resistant to degradation. 293T cells were transfected with the combination of vectors shown at the top. Cell lysates were analyzed by immunoblotting with antibodies against a FLAG epitope, COP1, an HA epitope, and γ-tubulin (E). Relative amounts of proteins were quantified using ImageJ software (NIH) (F).