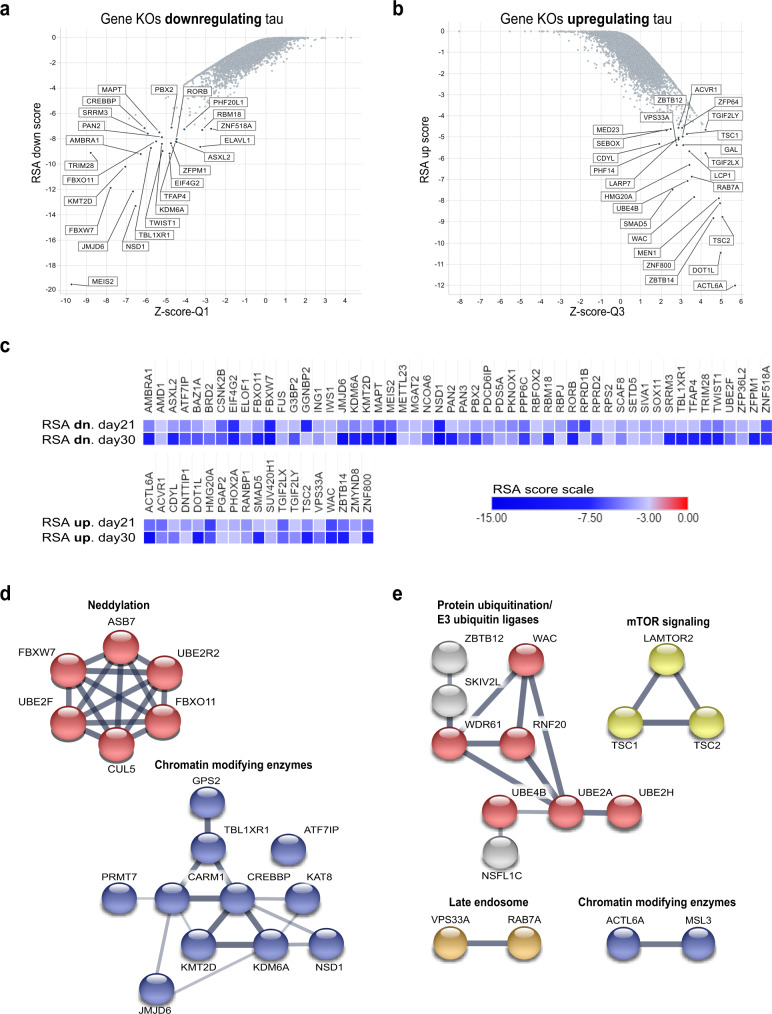
Fig. 2. Gene knockouts upregulating and downregulating tau protein in SH-SY5Y cells after CRISPR editing.
a, b Scatter plots with RSA scores (Y-axis) vs. z-score quartiles (X-axis), for 30 day edited group from primary genome-wide screen. Plots shows genes downregulating (a) and upregulating (b) tau protein levels. Highly significant candidate genes are labeled. c Heat maps representing candidate genes with an RSA score ≤−3.0 in primary genome-wide screens (21 and 30 day time points). d, e STRING enrichment pathways for highly validated genes (primary and mini array screens). Links between nodes are highly confident connections. d Pathways identified for genes downregulating tau protein levels. e Pathways identified for genes upregulating tau protein levels. KO knockout, Q quartile, Dn down, RSA redundant siRNA activity score.