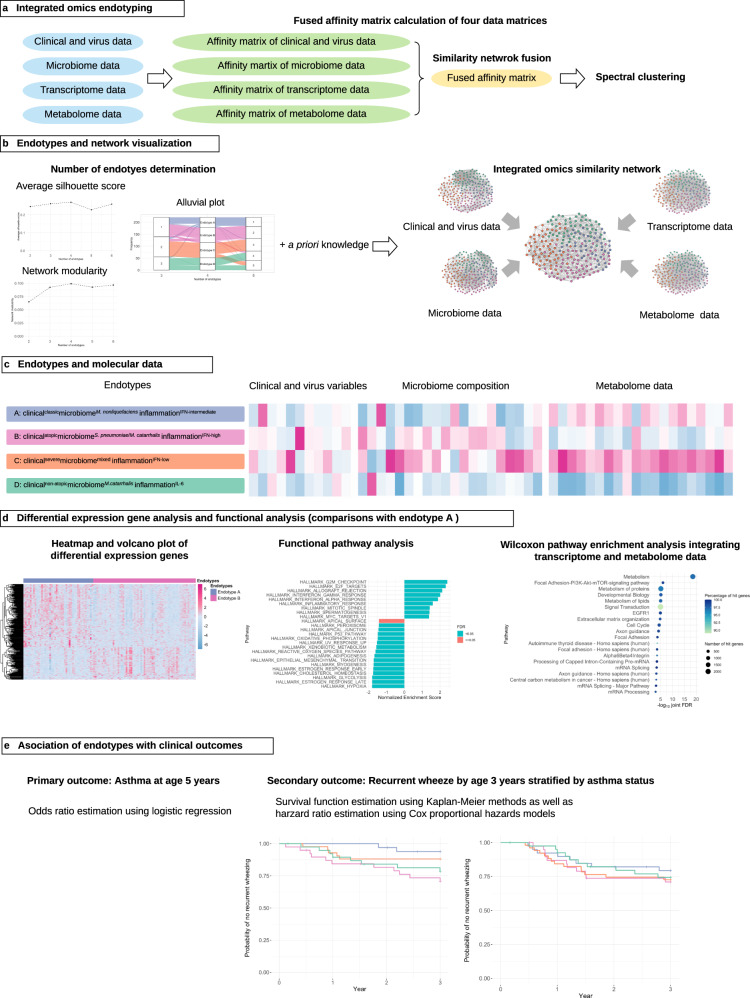
Fig. 1. Analytic workflow of integrated omics endotyping.
a After an affinity matrix of each dataset (clinical and virus, microbiome, transcriptome, and metabolome) was separately computed, and a fused affinity matrix was generated by similarity network fusion. Then, the fused affinity matrix was used to identify mutually exclusive endotypes by spectral clustering. b A combination of average silhouette scores, network modularity, and clinical plausibility (in addition to endotype size) was used to choose the optimal number of endotypes. The concordance between the different numbers of endotypes was also examined. After deriving endotypes, a similarity network was visualized. c Between four derived endotypes of RSV bronchiolitis, the differences in the major clinical and virus variables, nasopharyngeal microbiome, and metabolome were visualized using heatmap. d Differentially expressed genes (endotype A as the reference group) were visualized using a heatmap and volcano plot. The functional pathway analysis using the gene set enrichment analysis and the Wilcoxon pathway enrichment analysis integrating transcriptomic and metabolome data were conducted to identify enriched pathways. e The risk of childhood asthma (binary outcome) was modeled by fitting a logistic regression model. The rate of recurrent wheeze (time-to-event outcome) was modeled by fitting a Cox proportional hazards model. RSV respiratory syncytial virus, IFN interferon, IL interleukin.