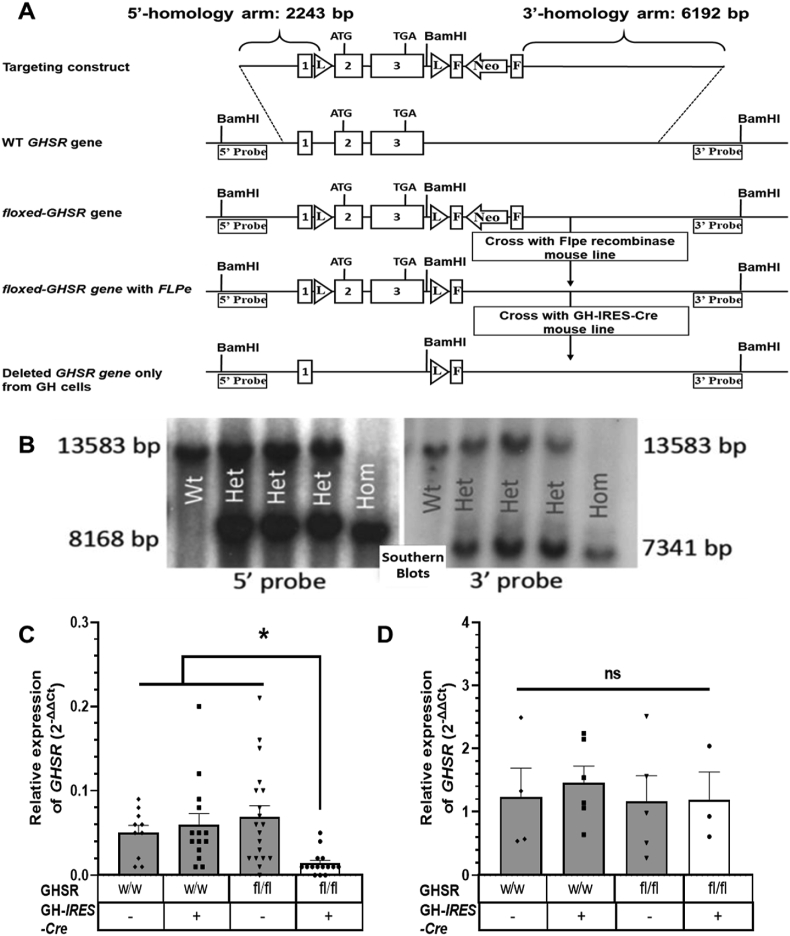
Figure 2.
Generation and validation of a novel floxed GHSR mouse line and mice with somatotroph-selective GHSR deletion. (A) Schematic diagrams showing the homologous recombination targeting strategy to flank the GHSR-coding region with loxP sites, creating floxed GHSR mice. Binding sites for the 5′ and 3′ Southern blotting probes used to detect correctly targeted genomic DNA (after restriction digestion with BamH1) are depicted. (B) Southern blotting of genomic DNA of representative pups derived from crosses of mice heterozygous for the loxP-flanked GHSR allele, including pups homozygous for the GHSRfl allele (GHSRfl/fl; “Hom”), mice with two copies of the wild-type GHSR allele (GHSRWT/WT; “Wt”), and heterozygotes (GHSRWT/fl; “Het”). Expression pattern of GHSR mRNA in pituitaries (C) and hypothalami (D) of mice generated by crossing GH-IRES-Cre mice to floxed GHSR mice. Data are relative 2−ΔΔCt values and were analyzed by one-way ANOVA followed by Sidak's multiple comparisons test. n = 10–19 (pituitary) and n = 3–6 (hypothalamus). All of the values are expressed as mean ± SEM. ∗P < 0.05 and ns = not significant. w/w, mice with 2 copies of the Wt GHSR gene; fl/fl, mice homozygous for the floxed GHSR gene; –, absence or +, presence of the GH-IRES-Cre allele.