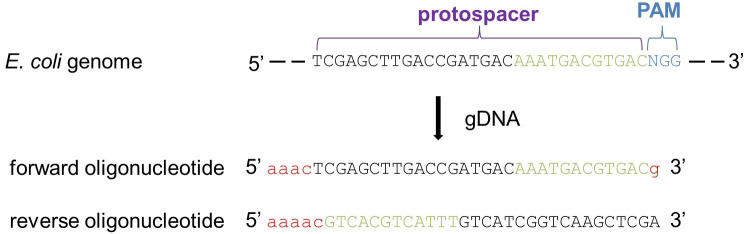
Figure 3. Illustration on how to select a gene-specific Cas9 target site.
To design the guide DNA (gDNA) identify a PAM sequence (5’-NGG-3’, where N stands for any of the four nucleotides) near the 5’ end of the target gene. Then, select the 30 nucleotides upstream of the PAM region (protospacer). BLAST the entire sequence (protospacer + PAM), which is going to be the core of the forward oligonucleotide of your gDNA, against the genome of the target organism. To avoid Cas9 off-target effect, a ‘reliable’ gDNA should not have any homology among the 10-12 PAM-proximal nucleotides (seed region, in green) and less than 15% homology among the remaining protospacer when blasted against the rest of the genome. Finally, the chosen protospacer + PAM sequence is completed by the addition of five base pairs (in red) to facilitate the ligation for the subsequent cloning of the pCRISPR-SacB-gDNA, resulting in the forward oligonucleotide of the gDNA sequence. The reverse oligonucleotide of the gDNA is the reverse complement of the protospacer + PAM forward sequence with the addition of five base pairs (in red) as done for the forward oligonucleotide.