Abstract
Alkaline phosphatase (ALP) is an important enzyme-based biomarker present in several bacterial species; however, it is currently undervalued as a strategy to detect pathogenic bacteria. Here, we explore our ALP-responsive colorimetric and fluorescent probe (TCF-ALP) for such applications. TCF-ALP displayed a colorimetric and fluorescence response towards Staphylococcus aureus (S. aureus), with a limit of detection of 3.7 × 106 CFU mL−1 after 24 h incubation. To our surprise, TCF-ALP proved selective towards Staphylococcus bacteria when compared with Enterococcus faecalis (E. faecalis), and Gram-negative P. aeruginosa and E. coli. Selectivity was also seen in clinically relevant S. aureus biofilms. Owing to the high prevalence and surface location of S. aureus in chronic wounds, TCF-ALP was subsequently encapsulated in polyvinyl alcohol (PVA)-based hydrogels as a proof-of-concept “smart” wound dressing. TCF-ALP hydrogels were capable of detecting S. aureus in planktonic and biofilm assays, and displayed a clear colour change from yellow to purple after 24 h incubation using ex vivo porcine skin models. Overall, TCF-ALP is a simple tool that requires no prior knowledge, training, or specialist equipment, and has the potential to overcome issues related to invasive swabbing and tissue biopsy methods. Thus, TCF-ALP could be used as a tool to monitor the early development of infection in a wound and allow for the rapid provision of appropriate treatment for Staphylococcal bacterial infections.
The optical response of an alkaline phosphatase (ALP) probe was evaluated against various bacterial species.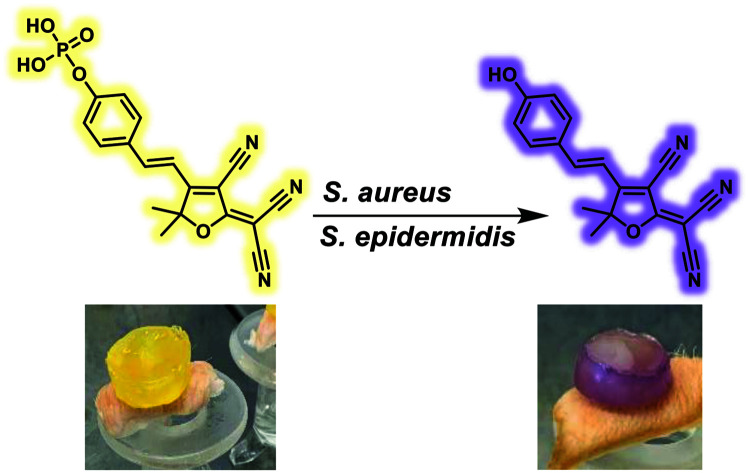
Introduction
Chronic wounds affect the lives of millions worldwide and exert significant financial pressure on healthcare systems, with the NHS spending an estimated £ 4–5 billion per year.1,2 These non-healing wounds can persist for prolonged periods of time (months to years), often affecting patients with underlying health conditions such as diabetes, obesity, and cancer.1,2 Bacterial infections contribute significantly to the non-healing nature of these wounds with bacteria often forming complex bacterial communities known as biofilms.3 Protected by extracellular polymeric substances (EPS), biofilms result in hard-to-treat infections, abet in the development of antimicrobial resistance, and abet inflammation at the wound site, resulting in further tissue damage.6,7 If left untreated, these localised infections can develop into life-threatening systemic infections.8,9 As a result, effective wound management combined with the ability to rapidly identify and treat pathogenic bacteria is highly desirable for the treatment of chronic wounds.
Swab and tissue biopsy methods are currently the gold standard used to confirm the presence of a wound infection and used to identify the infection-causing pathogenic bacteria.10,11 However, these invasive and painful techniques have inherent limitations, requiring time-consuming protocols, highly trained specialists, and are limited to bacterial species that can be routinely grown in a laboratory.10,12 These limitations result in delayed treatment, potentially leading to a worse prognosis for the patient.4 Recent technological advancements have led to the development of assays that utilise genomic markers to rapidly identify pathogenic bacteria (ELISA, PCR, DNA microarrays and optical and electrochemical biosensors).5–7,8–12 However, as with conventional methods, most of these techniques are expensive and require extensive sample manipulation by trained specialists.13–15
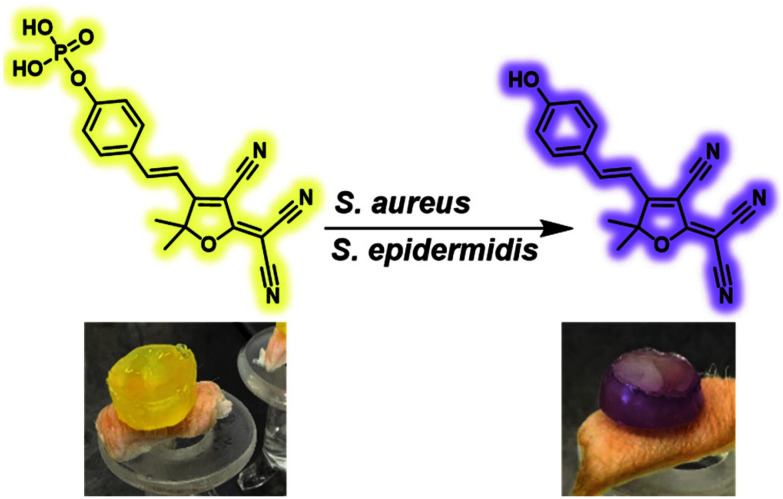
Small molecule fluorescent and colorimetric probes provide an attractive alternative as they enable non-specialist use, are easy to handle and store, and confer a high degree of selectivity and sensitivity towards bacterial-based biomarkers.16–22 In addition, they offer a complementary strategy to smart wound technologies and point of care (PoC) devices.23,24 Amongst the bacterial based biomarkers, alkaline phosphatase (ALP) is an enzyme responsible for the hydrolysis of phosphoesters to facilitate the release of inorganic phosphate (Pi), an essential nutrient for bacterial cell growth.25 ALP is present in numerous bacteria species, including the ESKAPE pathogens, Escherichia coli (E. coli),26Pseudomonas aeruginosa (P. aeruginosa),27 and Staphylococcus aureus (S. aureus).28 Therefore, it is not surprising that the determination of ALP activity has been a focal point for numerous microbiological studies,29,30 and as a strategy for the development of ALP-responsive small-molecule probes for the in vitro detection of bacteria.31–33 However, in comparison to other enzyme-based bacterial biomarkers, this enzyme has been undervalued as a strategy to detect pathogenic bacteria using small molecule fluorescent probes. In addition, recent studies have failed to develop viable platforms with the potential for clinical applications. In this work, we evaluated the ability of our previously designed ALP responsive fluorescent probe TCF-ALP to detect pathogenic bacteria (Scheme 1).34TCF-ALP was shown to selectively detect planktonic S. aureus and Staphylococcus epidermidis (S. epidermidis), and was effective in detecting S. aureus in a series of planktonic, biofilm, and ex vivo studies. Our work clearly demonstrates the use of TCF-ALP as a simple tool that could be used to monitor the early development of infection and allow for the rapid provision of the appropriate treatment for Staphylococcal bacterial infections. It is important to note that in this study, only clinically relevant Staphylococcus species (S. aureus and S. epidermidis35) were used with TCF-ALP.
Scheme 1. The use of TCF-ALP for the selective detection of Staphylococcus spp. via alkaline phosphatase dephosphorylation. Images show an ex vivo experiment utilising a PVA-hydrogel encapsulated with TCF-ALP before and after S. aureus NCTC 10788 inoculation.
Results and discussion
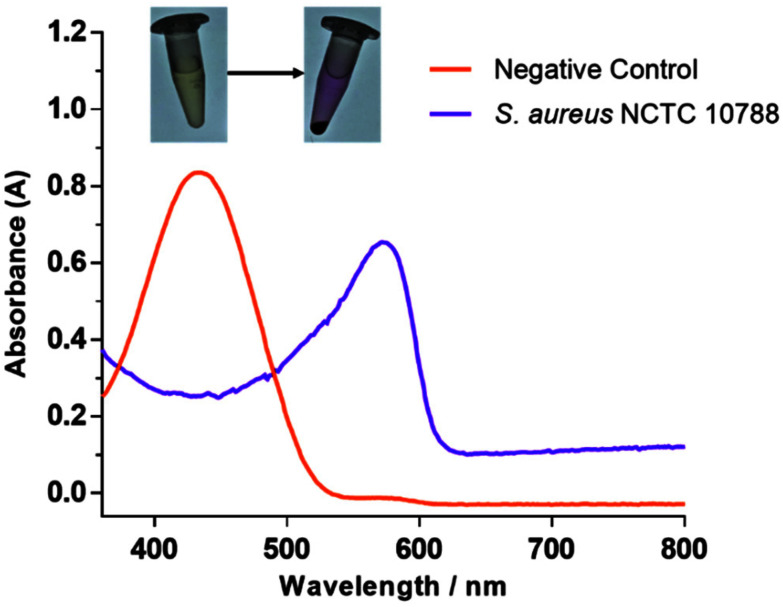
Gram-positive S. aureus is often one of the first bacteria to establish within a wound, primarily colonising close to the surface.36 In comparison, Gram-negative P. aeruginosa is a late-stage coloniser of chronic wounds and is predominately found in the deeper regions of the wound.36 Therefore, the surface location and high prevalence of S. aureus represents a unique opportunity to be used for the early diagnosis of chronic wound development. Hence, with this work, S. aureus NCTC 10788 was chosen for evaluating the ability of TCF-ALP as a diagnostic tool. Initial experiments revealed that incubation of TCF-ALP (10 μM) with S. aureus NCTC 10788 (108 CFU mL−1) for 24 h led to a bathochromic shift in UV-Vis absorption and an easy-to-visualise colour change from yellow to purple (Fig. 1). This result was indicative of the ALP-mediated transformation of TCF-ALP to TCF-OH.
Fig. 1. UV-Vis spectra of TCF-ALP (10 μM) after 24 h incubation at 32 °C with S. aureus NCTC 10788 (108 CFU mL−1) in 50 mM Tris-HCl buffer, pH = 9.2.
For all experiments, S. aureus NCTC 10788 was grown for 24 h at 32 °C in Mueller Hinton broth (MH; pH 7.4) before centrifugation and resuspension in a solution of TCF-ALP (10 μM, 50 mM Tris HCl, pH 9.2). The resuspended solution was held at pH of 9.2 since this previously has been shown to be optimum for ALP activity.34 MH broth was chosen over other non-differential broths such as Tryptic Soy Broth (TSB) and Luria–Bertani (LB) as it provided the greatest fluorescence response (Fig. S1 and S2†). The difference in response was attributed to the varying amounts of inorganic phosphate (Pi) found in each medium, which can serve as an ALP inhibitor.37 Optimisation experiments ensured no interference or unwanted response occurred from residual growth medium (S. aureus bacterial pellets were washed 1 to 3 times with Tris-HCl buffer [50 mM, pH 9.2] – Fig. S3†).
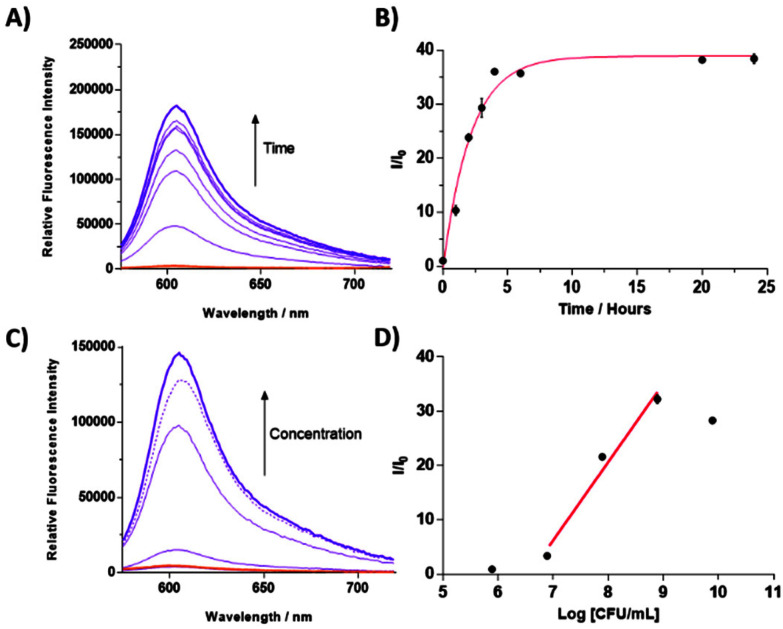
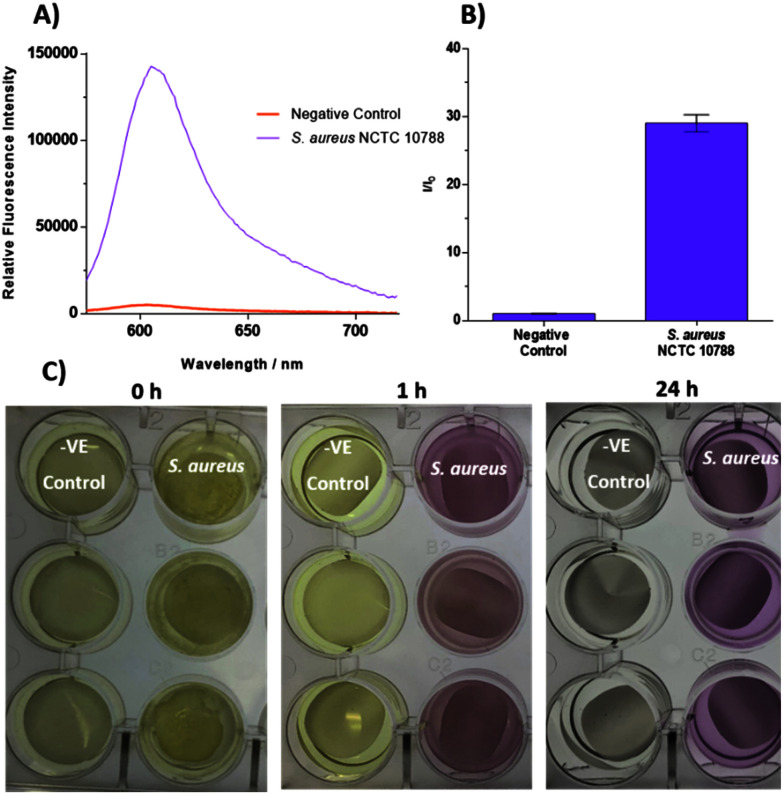
Given that our overall goal was to use TCF-ALP in wound dressing/diagnostic applications, we investigated the overall fluorescence response of TCF-ALP towards S. aureus under varying incubation temperatures (25 °C – room temperature, 32 °C – surface temperature of skin and 37 °C – optimum growth conditions) (Fig. S4†). Marginal differences were observed for the overall fluorescence responses when TCF-ALP was incubated with S. aureus at each of these temperatures, demonstrating the suitability of the system for use as a PoC device or in smart wound applications. Following on from our initial studies, fluorescence analysis was performed to determine the minimum incubation time needed for S. aureus NCTC 10788 (108 CFU mL−1) to generate a response (Fig. 2A and B). After 1 h incubation, an approximate 10-fold increase in fluorescence intensity was observed, which continued to increase to a >30-fold increase in fluorescence intensity after approximately 10 h.
Fig. 2. (A) Fluorescence spectra of TCF-ALP (10 μM) recorded over the course of 24 h upon addition of S. aureus NCTC 10788 (108 CFU mL−1) in 50 mM Tris-HCl buffer pH = 9.2 at 32 °C. (B) Corresponding change in fluorescence (I/I0) of TCF-ALP (10 μM) upon addition of S. aureus NCTC 10788 (108 CFU mL−1) in 50 mM Tris-HCl buffer pH = 9.2 at 32 °C. Error bars indicate standard deviation (n = 3). (C) Fluorescence spectra of TCF-ALP (10 μM) after 24 h incubation with various concentrations of S. aureus NCTC 10788 (0–109 CFU mL−1) in 50 mM Tris-HCl buffer pH = 9.2 at 32 °C with the dotted line representing 1010 CFU mL−1, and (D) corresponding sensitivity graph. (X-intercept 6.568 = 3.70 × 106 CFU mL−1; Y = 14.45X–94.88; R2 0.9759). All experiments used λex = 542 (bandwidth 15) nm and λem = 606 nm.
Next, TCF-ALP (10 μM) was incubated with varying concentrations of S. aureus NCTC 10788 (105–1010 CFU mL−1), and the fluorescence intensity was recorded after 1 h and 24 h incubation. After 1 h incubation, a linear fluorescence increase was observed for bacterial concentrations between 8–10 log CFU mL−1 (Fig. S5†). The LOD, as determined by linear regression, was found to be 3.17 × 107 CFU mL−1 (95% confidence interval (CI) 2.95–3.40 × 107 CFU mL−1). After 24 h incubation, a linear fluorescence increase was observed for bacterial concentrations between 7–9 log CFU mL−1 (Fig. 2C and D). A slight decline in the fluorescence response was observed for 1010 CFU mL−1 of S. aureus NCTC 10788; this is believed to be the result of the cellular uptake of TCF-ALP at high bacterial concentrations. This hypothesis was supported through fluorescence imaging studies (see ESI – Fig. S6 and S7†). At 24 h, the LOD was determined as 3.7 × 106 CFU mL−1 (95% CI of 2.08–5.79 × 106 CFU mL−1). This LOD is in line with other fluorescent and colorimetric probes used for the detection of a variety of bacterial enzymes (Table S2†). Biofilms are known to have a significantly higher bacterial cell density compared to the concentration of planktonic bacteria thought to result in a localised infection (105 CFU mL−1).38 As a result, we believe the high LOD of TCF-ALP is advantageous, as the clear “turn-on” fluorescence and colorimetric response is witnessed at infection-related concentrations (>106 CFU mL−1). Fluorescent/colorimetric probes with a LOD below these values may lead to false positive results (Table S2†).
Bacterial selectivity of TCF-ALP
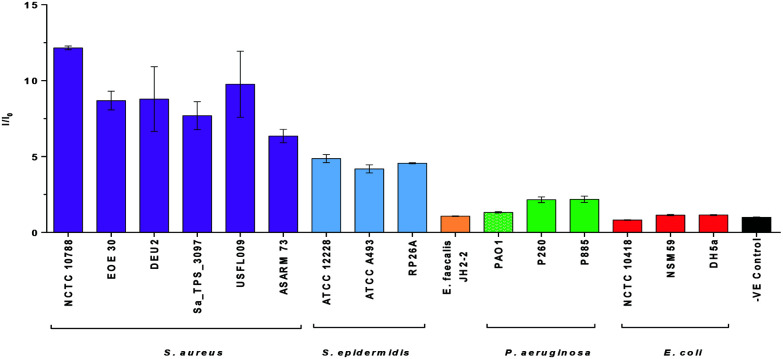
Diagnostic devices that selectively detect specific types of bacteria are particularly attractive as they would allow for the rapid provision of the appropriate treatment to improve prognosis and avoid the misuse of antibiotics. This led us to perform selectivity studies for TCF-ALP against various bacterial species. In this study, we initially evaluated the fluorescence response of TCF-ALP against six Gram-positive S. aureus strains, three S. epidermidis strains, one Gram-positive Enterococcus faecalis (E. faecalis) strain, and three bacterial strains of Gram-negative P. aeruginosa and E. coli. All isolates used in this study were either relevant clinical isolates or reference strains, with the province outlined in the ESI – see Methods (Table S1†). As seen in Fig. 3, all Staphylococcal strains displayed a clear fluorescence turn-on response. However, Gram-positive E. faecalis and all Gram-negative bacteria tested resulted in a negligible increase in fluorescence intensity, even after 24 h (see ESI – Fig. S8†). Cell counting confirmed that TCF-ALP had a minimal effect on Gram-negative bacterial cell viability (Fig. S9†). Additionally, the commercially available colorimetric ALP probe, p-nitrophenyl phosphate (p-NPP), was used to determine ALP activity in each bacterial species. While S. aureus, E. faecalis and E. coli were shown to have similar ALP activities, minimal ALP activity was observed for P. aeruginosa (Fig. S10 and S11†). These experiments further suggested the selectivity of TCF-ALP towards Staphylococcal species. To further illustrate this selectivity, TCF-ALP was then evaluated against a total of 42 S. aureus isolates. Remarkably, all S. aureus strains produced at least a 10-fold increase in fluorescence intensity after 24 h, regardless of the phenotype (Fig. S12 and S13†). This demonstrates the potential use of TCF-ALP as a tool for the rapid detection of S. aureus in a clinical setting.
Fig. 3. Selectivity bar chart of TCF-ALP (10 μM) in 50 mM Tris-HCl buffer pH = 9.2 after 1 h incubation with various bacterial strains (108 CFU mL−1) at 32 °C. λex = 542 (bandwidth 15) nm. λem = 606 nm. Error bars indicate standard deviation (n = 3).
Detection of ALP within S. aureus biofilms
As bacterial biofilms are present in the majority of infected chronic wounds,6,7 we turned our attention towards the utility of TCF-ALP to respond to S. aureus biofilms. Remarkably, when 24 h old S. aureus NCTC 10788 biofilms (formed in 96-well microtiter plates) were incubated with TCF-ALP (10 μM) for 24 h at 32 °C, we observed up to a 40-fold increase in fluorescence intensity (Fig. S14†). ALP inhibition experiments were performed to ensure that ALP activity was responsible for the fluorescence “turn-on” of TCF-ALP. Upon increasing concentrations of a known inhibitor (Sodium orthovanadate) a decrease in fluorescence intensity was observed, confirming the ALP-mediated hydrolysis of TCF-ALP (Fig. S15 and S16†).
With these promising results in hand, a more robust biofilm model was used, termed the colony biofilm model; this provides a more accurate representation of a biofilm in a clinical setting.39 Colony biofilms were grown on a permeable polycarbonate membrane on a MH agar plate – used to mimic the surface of an infection site. A representative wound environment was achieved by treating the polycarbonate membranes with artificial wound fluid (AWF; 50% fetal bovine serum in 50% peptone water [0.9% sodium chloride in 0.1% peptone]) before the addition of bacteria. As seen in Fig. 4A and B, a large increase in fluorescence intensity was observed when S. aureus NCTC 10788 biofilms were incubated with TCF-ALP (10 μM), compared to the negative control. A positive turn-on response and a colour change from yellow to purple also occurred after only 1 h incubation with TCF-ALP (Fig. 4C and S17†). Conversely, minimal fluorescence responses were observed when TCF-ALP was incubated with Gram negative bacterial biofilms, further highlighting the selectivity of this probe (Fig. S18–S20†) TCF-ALP also had minimal effect on the bacterial viability of all bacterial biofilms tested (Fig. S21†).
Fig. 4. (A) Fluorescence spectra of TCF-ALP (10 μM) after 24 h incubation with biofilms of S. aureus NCTC 10788 (1011 CFU per membrane) in 50 mM Tris-HCl buffer pH = 9.2 at 32 °C. (B) Corresponding selectivity bar chart. λex = 542 (bandwidth 15) nm. λem = 606 nm. Error bars indicate standard deviation (n = 3). (C) Images taken of negative controls (Membrane and Artificial Wound Fluid (AWF) only) and biofilms of S. aureus NCTC 10788 (1011 CFU per membrane) after 0, 1, and 24 h incubation with 10 μM TCF-ALP in 50 mM Tris-HCl buffer pH = 9.2 at 32 °C.
Incorporation of TCF-ALP into polyvinyl alcohol-based (PVA) hydrogels and use in the detection of S. aureus
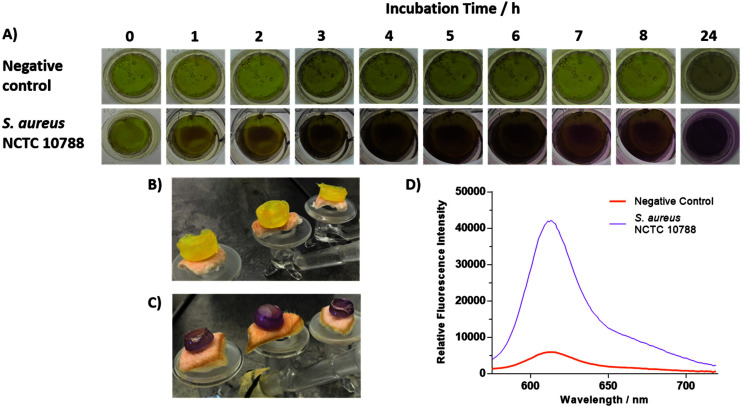
As previously mentioned, traditional methods of bacterial detection utilise swabbing and tissue biopsy. A major drawback to these techniques is the requirement for the removal of a conventional wound dressing. This has the potential to damage healing tissue, can cause pain to the patient, and risk exposure to infection.40 Hydrogels are being increasingly used in the field of wound care as they maintain a moist wound environment, accelerate the healing processes, are easy to remove, and are easy to develop and handle.41 However, only a few diagnostic hydrogel systems have currently been designed for the detection of bacteria.26,29 To demonstrate the potential of TCF-ALP for use in “smart” wound dressing systems,42 we turned our attention to the encapsulation of TCF-ALP within a PVA-based hydrogel. In brief, TCF-ALP (38.5 μL, 100 μM in DMSO) was suspended in 1 mL of 10% w/v PVA (50 mM Tris HCl, pH 9.2). This solution was then subjected to a single freeze–thaw cycle, producing a mechanically stable hydrogel. To account for any potential loss of probe during the freeze–thaw cycle, TCF-ALP was loaded into the hydrogels at a higher concentration (100 μM). With TCF-ALP encapsulated hydrogels in hand, we evaluated their ability to respond to planktonic S. aureus NCTC 10788. Each hydrogel was incubated in 2 mL of a 108 CFU mL−1S. aureus suspension in 50 mM Tris HCl (pH 9.2) and photographed at regular time intervals (Fig. S22†). After 5 h, the hydrogels exhibited a significant colour change from yellow to purple, progressing to a deep purple after 24 h incubation, and a strong corresponding fluorescence intensity at 606 nm — indicative of the formation of TCF-OH (Fig. S23†). While it is important to note that small amounts of TCF-ALP leached from the system, this is a proof-of-concept study with the opportunity to optimise the system at a later stage of development. Experiments with TCF-ALP-based hydrogels using S. aureus NCTC 10788 colony biofilms displayed a remarkable colour change from yellow to purple (Fig. 5A), with a concomitant increase in fluorescence intensity at 606 nm (Fig. S24†). Interestingly, the location on the hydrogel in which the change in colour gradually occurred was the contact point between the hydrogel and bacterial surface. Additionally, both the blank hydrogel and TCF-ALP hydrogel had no overall effect on the viability of S. aureus NCTC 10788 (Fig. S25†).
Fig. 5. (A) Images depicting the colour of TCF-ALP-based PVA hydrogels in the presence of a negative control (Membrane and Artificial Wound Fluid (AWF) only) and a S. aureus NCTC 10788 biofilm (1011 CFU per membrane). Images taken hourly at 0–8 h and 24 h and were repeated in triplicate. (B) TCF-ALP-based PVA hydrogels on non-inoculated porcine skin wound model after 24 h incubation. (C) TCF-ALP-based PVA hydrogels on a porcine skin wound model inoculated with S. aureus NCTC 10788 (10 μL of 108 CFU mL−1), and (D) corresponding fluorescence intensities of the hydrogels shown in (B) and (C). λex = 542 (bandwidth 15) nm. λem = 606 nm.
Ex vivo porcine skin models
Finally, to demonstrate the potential diagnostic capability of TCF-ALP, ex vivo skin models were performed using porcine skin, owing to its similarity to human skin.43 In this ex vivo study, porcine skin was treated with 10 μL of S. aureus NCTC 10788 (108 CFU mL−1) and allowed to dry at room temperature. Subsequent 24 h incubation of the S. aureus-treated porcine skin with 1 mL of TCF-ALP (10 μM. 50 mM Tris HCL, pH 9.2) resulted in an 8-fold increase in fluorescence intensity, with an associated colour change from yellow to purple; no overall change to cell concentration was observed (Fig. S26–S28†). Additionally, TCF-ALP hydrogels were evaluated using the ex vivo model in an attempt to model a clinically relevant situation. To our excitement, TCF-ALP hydrogels placed on S. aureus-treated porcine skin resulted in a clear colour change from yellow to purple after 24 h incubation. In addition, this colour change corresponded to an approximate 7-fold increase in fluorescence intensity (Fig. 5B–D), with no difference in bacterial concentration observed (Fig. S29†). As such, we believe these results demonstrate the potential of TCF-ALP to be used in the development of smart wound dressings for the rapid identification of an infected wound to provide the appropriate treatment without removal of the patient's wound dressing.
Conclusion
Rapid detection of bacterial species present within a wound is important for the development of suitable and rapid treatment protocols. New methods that allow for quick PoC detection are thus of great importance, especially as we continue to enter the era of antibiotic resistance. With this aim in mind, the ALP-responsive colorimetric and fluorescent probe TCF-ALP was evaluated for its response to pathogenic bacteria. In the presence of Gram-positive S. aureus NCTC 10788, TCF-ALP was shown to have an excellent colorimetric and fluorescence response with a limit of detection of 3.7 × 106 CFU mL−1 after 24 h incubation. To our surprise and delight, TCF-ALP proved selective towards S. aureus and S. epidermidis compared to one Gram-positive E. faecalis strain and six Gram-negative strains (three P. aeruginosa and three E. coli) with selectivity seen in clinically relevant biofilm models. TCF-ALP was then evaluated against a total of 42 S. aureus isolates. Remarkably, all 42 S. aureus strains produced at least a 10-fold increase in fluorescence intensity after 24 h, regardless of the phenotype. TCF-ALP was encapsulated in PVA-based hydrogels as a proof of concept for “smart” wound dressing applications. TCF-ALP hydrogels proved effective for the detection of S. aureus as planktonic and biofilm bacteria. Ex vivo skin models with TCF-ALP hydrogels resulted in a clear colour change from yellow to purple after 24 h incubation. Overall, TCF-ALP represents a simple diagnostic tool that requires no prior knowledge, no training/specialist equipment, and overcomes the time consuming and invasive swabbing and tissue biopsy methods. Thus, TCF-ALP could be used as a tool to monitor the early development of infection in a wound and allow the rapid provision of the appropriate treatment for Staphylococcal bacterial infections.
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
This work was supported in part by grant MR/N0137941/1 for the GW4 BIOMED DTP, awarded to the Universities of Bath, Bristol, Cardiff and Exeter from the Medical Research Council (MRC)/UKRI. The authors thank the National Natural Science Foundation of China (No. 21788102, 91853201, 21722801, 81673489 and 31871414), the Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX03), the International Cooperation Program of Shanghai Science and Technology Committee (No. 17520750100) and the Fundamental Research Funds for the Central Universities (222201717003) for financial support. LG and ATAJ would like to thank Dr Maisem Laabei for his help in acquiring S. aureus strains for testing. GW would like to thank the GCDC at the University of Kent for funding. The authors would like to thank Kaya Davies-Brenchley for her help in the preparation of this manuscript. ACS would like to extend his thanks to Sajal Sen (The University of Texas at Austin) and James T. Brewster (Harvard University) for helpful discussions. TDJ wishes to thank the Royal Society for a Wolfson Research Merit Award and the Open Research Fund of the School of Chemistry and Chemical Engineering, Henan Normal University for support (2020ZD01).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0bm01918f
References
- Falanga V. Lancet. 2005;366:1736–1743. doi: 10.1016/S0140-6736(05)67700-8. [DOI] [PubMed] [Google Scholar]
- Sen C. K. Adv. Wound Care. 2019;8:39–48. doi: 10.1089/wound.2019.0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H. C. Wingender J. Szewzyk U. Steinberg P. Rice S. A. Kjelleberg S. Nat. Rev. Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- Haalboom M. Blokhuis-Arkes M. H. E. Beuk R. J. Meerwaldt R. Klont R. Schijffelen M. J. Bowler P. B. Burnet M. Sigl E. van der Palen J. A. M. Clin. Microbiol. Infect. 2019;25 doi: 10.1016/j.cmi.2018.08.012. [DOI] [PubMed] [Google Scholar]
- Law J. W. F. Ab Mutalib N. S. Chan K. G. Lee L. H. Front. Microbiol. 2015;5 doi: 10.3389/fmicb.2014.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N. Bhardwaj S. K. Nayak M. K. Mehta J. Kim K. H. Deep A. TrAC, Trends Anal. Chem. 2017;97:120–135. [Google Scholar]
- Richardson M. D. Warnock D. W. Sabouraudia. 1983;21:1–14. doi: 10.1080/00362178385380031. [DOI] [PubMed] [Google Scholar]
- Zhou B. L. Xiao J. W. Liu S. F. Yang J. Wang Y. Nie F. P. Zhou Q. Li Y. G. Zhao G. H. Food Control. 2013;32:198–204. [Google Scholar]
- Wei S. Daliri E. B. M. Chelliah R. Park B. J. Lim J. S. Baek M. A. Nam Y. S. Seo K. H. Jin Y. G. Oh D. H. J. Food Saf. 2019;39 doi: 10.1111/jfs.12558. [DOI] [Google Scholar]
- Yang Y. Y. Rajendran V. Jayaraman V. Wang T. H. Bei K. Krishna K. Rajasekaran K. Rajasekaran J. J. Krishnamurthy H. Gut Pathog. 2019;11 doi: 10.1186/s13099-019-0329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Y. Liu D. Cao B. Y. Han W. Q. Liu Y. Q. Liu F. X. Guo X. Bastin D. A. Feng L. Wang L. J. Clin. Microbiol. 2006;44:4376–4383. doi: 10.1128/JCM.01389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. M. Lee S. Y. Trends Biotechnol. 2016;34:7–25. doi: 10.1016/j.tibtech.2015.09.012. [DOI] [PubMed] [Google Scholar]
- Zhang P. Liu H. Ma S. Z. Men S. Li Q. Z. Yang X. Wang H. N. Zhang A. Y. Biosens. Bioelectron. 2016;80:538–542. doi: 10.1016/j.bios.2016.02.031. [DOI] [PubMed] [Google Scholar]
- Liu X. Hu Y. X. Zheng S. Liu Y. He Z. Luo F. Sens. Actuators, B. 2016;230:191–198. [Google Scholar]
- Alocilja E. C. Radke S. M. Biosens. Bioelectron. 2003;18:841–846. doi: 10.1016/s0956-5663(03)00009-5. [DOI] [PubMed] [Google Scholar]
- Gwynne L. Williams G. T. Yan K.-C. Gardiner J. E. Hilton K. L. F. Patenall B. L. Hiscock J. R. Maillard J.-Y. He X.-P. James T. D. Sedgwick A. C. Jenkins A. T. A. Isr. J. Chem. doi: 10.1002/ijch.202000105. [DOI] [Google Scholar]
- Sedgwick A. C. Brewster J. T. Wu T. H. Feng X. Bull S. D. Qian X. H. Sessler J. L. James T. D. Anslyn E. V. Sun X. L. Chem. Soc. Rev. 2021;50:9–38. doi: 10.1039/c9cs00538b. [DOI] [PubMed] [Google Scholar]
- Wu D. Sedgwick A. C. Gunnlaugsson T. Akkaya E. U. Yoon J. James T. D. Chem. Soc. Rev. 2017;46:7105–7123. doi: 10.1039/c7cs00240h. [DOI] [PubMed] [Google Scholar]
- Jia Z. Y. Gwynne L. Sedgwick A. C. Muller M. Williams G. T. Jenkins A. T. A. James T. D. Schonherr H. ACS Appl. Bio Mater. 2020;3:4398–4407. doi: 10.1021/acsabm.0c00403. [DOI] [PubMed] [Google Scholar]
- Ebrahimi M. M. S. Voss Y. Schonherr H. ACS Appl. Mater. Interfaces. 2015;7:20190–20199. doi: 10.1021/acsami.5b05746. [DOI] [PubMed] [Google Scholar]
- Kocaoglu O. Carlson E. E. Nat. Chem. Biol. 2016;12:472–478. doi: 10.1038/nchembio.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchido Y. Horiuchi R. Hashimoto T. Ishihara K. Kanzawa N. Hayashita T. Anal. Chem. 2019;91:3929–3935. doi: 10.1021/acs.analchem.8b04870. [DOI] [PubMed] [Google Scholar]
- Brown M. S. Ashley B. Koh A. Front. Bioeng. Biotechnol. 2018;6 doi: 10.3389/fbioe.2018.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S. J. Kuan C. M. Hung M. W. Fu Y. Yeh J. A. Yao D. J. Cheng C. M. Micromachines. 2018;9 doi: 10.3390/mi9080418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajid A. Arora G. Singhal A. Kalia V. C. Singh Y. Annu. Rev. Microbiol. 2015;69:527–547. doi: 10.1146/annurev-micro-020415-111342. [DOI] [PubMed] [Google Scholar]
- Agrawal D. K. Wanner B. L. J. Bacteriol. 1990;172:3180–3190. doi: 10.1128/jb.172.6.3180-3190.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. J. Ingram J. M. Costerton J. W. J. Bacteriol. 1970;104:748–753. doi: 10.1128/jb.104.2.748-753.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D. B. Blobel H. J. Bacteriol. 1967;94:780–781. doi: 10.1128/jb.94.3.780-781.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. T. Xu K. D. McFeters G. A. Stewart P. S. Appl. Environ. Microbiol. 1998;64:1526–1531. doi: 10.1128/aem.64.4.1526-1531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M. Wasaki J. Tomizawa Y. Shinano T. Osaki M. Soil Sci. Plant Nutr. 2008;54:62–71. [Google Scholar]
- Sedgwick A. C. Yan K.-C. Mangel D. N. Shang Y. Steinbrueck A. Han H.-H. Brewster J. T. Hu X.-L. Snelson D. W. Lynch V. M. Tian H. He X.-P. Sessler J. L. J. Am. Chem. Soc. 2021;143:1278–1283. doi: 10.1021/jacs.0c11641. [DOI] [PubMed] [Google Scholar]
- Zhang X. Ren C. Hu F. Gao Y. Wang Z. Li H. Liu J. Liu B. Yang C. Anal. Chem. 2020;92:5185–5190. doi: 10.1021/acs.analchem.9b05704. [DOI] [PubMed] [Google Scholar]
- Kang E. B. Mazrad Z. A. I. Robby A. I. In I. Park S. Y. Eur. Polym. J. 2018;105:217–225. [Google Scholar]
- Gwynne L. Sedgwick A. C. Gardiner J. E. Williams G. T. Kim G. Lowe J. P. Maillard J. Y. Jenkins A. T. A. Bull S. D. Sessler J. L. Yoon J. James T. D. Front. Chem. 2019;7 doi: 10.3389/fchem.2019.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R. A. Rodvold K. A. Clin. Pharm. 1987;6:464–475. [PubMed] [Google Scholar]
- Fazli M. Bjarnsholt T. Kirketerp-Moller K. Jorgensen B. Andersen A. S. Krogfelt K. A. Givskov M. Tolker-Nielsen T. J. Clin. Microbiol. 2009;47:4084–4089. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbel S. Smirnov A. Chouayekh H. Sperandio B. Esnault C. Kormanec J. Virolle M. J. J. Bacteriol. 2006;188:6269–6276. doi: 10.1128/JB.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler P. G. Ostomy Wound Manag. 2003;49:44–53. [PubMed] [Google Scholar]
- Bahamondez-Canas T. F. Heersema L. A. Smyth H. D. C. Biomedicines. 2019;7 doi: 10.3390/biomedicines7020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhivya S. Padma V. V. Santhini E. Biomedicine. 2015;5:24–28. doi: 10.7603/s40681-015-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun E. A. Kenawy E.-R. S. Chen X. J. Adv. Res. 2017;8:217–233. doi: 10.1016/j.jare.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derakhshandeh H. Kashaf S. S. Aghabaglou F. Ghanavati I. O. Tamayol A. Trends Biotechnol. 2018;36:1259–1274. doi: 10.1016/j.tibtech.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T. P. Eaglstein W. H. Davis S. C. Mertz P. Wound Repair Regen. 2001;9:66–76. doi: 10.1046/j.1524-475x.2001.00066.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.