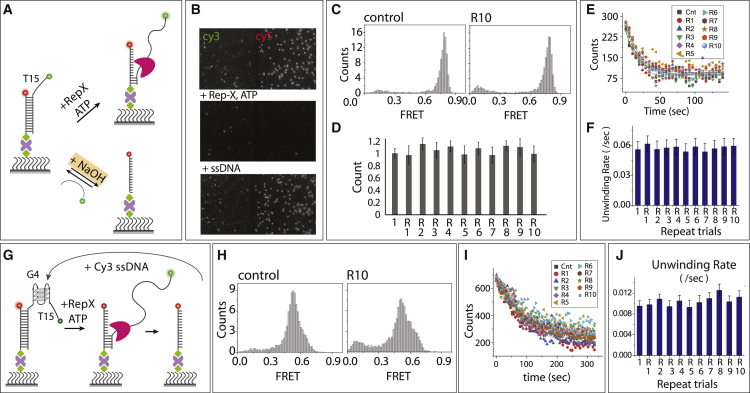
Figure 4.
Duplex recovery by denaturation and reannealing. (A) Schematic model of Rep-X-induced unwinding of partial duplex and reannealing by ssDNA. Low concentration of NaOH (∼50 mM) denatures the DNA duplex, which can be recovered by reannealing. (B) Representative fields of view before and after unwinding followed by reannealing. (C) The FRET histogram of control and after 10th reannealing. (D) The number of molecules counted during each repeat of reannealing and normalized with control. Error bar represents the SD from molecule counts of 20 different field of view. (E) Number of molecules counted for each repeated trial during unwinding of partial duplex and unwinding rate obtained from the single-exponential fitting (F). (G) Schematic model of Rep-X-induced G4 unwinding and reannealing by G4-ssDNA. (H) The FRET histogram of control and after 10th reannealing. (I) Number of molecules counted over time of G4 unwinding and the calculated unwinding rate (J). (F and J) Errors are reported from curve fitting. To see this figure in color, go online.