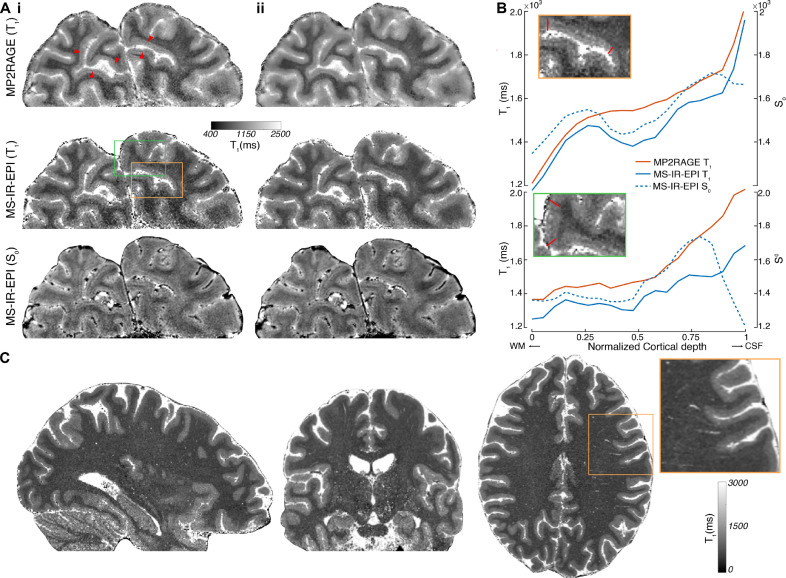
Fig. 8.
(Ai) High spatial resolution (0.35 × 0.35 × 0.7 mm3) T1 maps generated from MP2RAGE data (T11/T12=900/2375 ms, TRshot=4.5 s, 55 slices, 12 min 8 s acquisition time; top raw) and fitted T1 (middle raw) and S0 (bottom raw) maps derived from MS-IR-EPI data (TR=3.2 s, TE=20 ms, EPI factor=13, 35 slices, offsets=[0,7,14,21,28], 3 averages, 11 min 8 s total acquisition time) for an example slice of a partial brain dataset. The red arrowheads point to the Stria of Gennari. (ii) Corresponding maps after complex de-noising. (B) Cortical profiles for each of the original data sets (without de-noising) shown in (A) – mean across profiles between the two boundaries indicated by the red lines in the insets (top left) for a region within the striate cortex (top) and a region outside the striate cortex (bottom). Cortical profiles are plotted from the WM boundary to CSF boundary. Profiles from the striate cortex show a dip in T1 (and S0), corresponding to the Stria of Gennari, for the MS-IR-EPI data (solid and dashed blue profiles, respectively) but not for the MP2RAGE data (orange), whereas profiles outside the Stria do not show a dip. (C) Whole head MS-IR-EPI T1-data acquired at 0.5 mm isotropic resolution (EPI factor=7, TE=14 ms, TR=3.8 s, offsets=[0,8,16, 24, 32, 40, 48, 56, 64, 72], SMS=3, 240 slices, 18 min 44 s acquisition time). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)