Fig. 1.
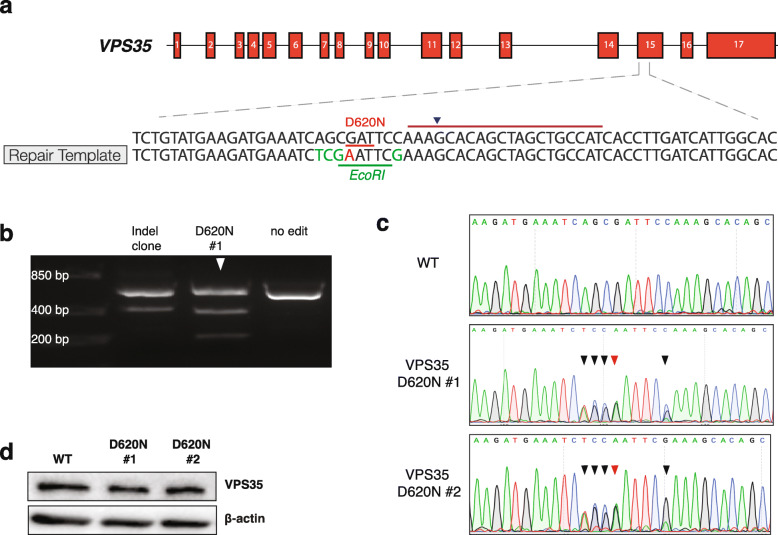
Generation of heterozygous VPS35D620N SH-SY5Y cells. a Schematic representation of cloning strategy for the generation of nucleotide exchange in the VPS35 locus that leads to the D620N mutation using CRISPR-Cas9 technology. Partial sequence of endogenous target on exon 15 is shown (upper sequence). The 20-nt guide sequence is depicted by the burgundy line with the blue arrowhead pointing towards the predicted Cas9-cleavage site. GAT sequence that encodes the aspartic acid residue at position 620 is highlighted in red. Lower sequence shows the design of the repair template used to create the mutation. The substitution G > A is shown in red. Silent substitutions to create an EcoRI restriction site are shown in green. b Representative agarose gel showing PCR products of the target VPS35 region obtained from different CRISPR-clones. PCR products were subjected to EcoRI digestion before loading on an agarose gel. Upper bands show noncleaved bands of the PCR amplicon. Arrowhead indicates an example of a successfully edited clone (VPS35D620N clone 1) that was partially cleaved by EcoRI. c Sanger chromatograms of the region surrounding the CRISPR-edit of WT (upper panel), VPS35 D620N clone 1 (middle panel) and VPS35 D620N clone 2 (lower panel). Red arrowhead indicates successful integration of the PD-associated mutation G > A. Black arrowheads indicate silent substitutions to create the EcoRI restriction site. Note that double peaks are shown for the edited nucleotides as one allele remains unaltered. d Representative immunoblots of protein extracts from WT and VPS35D620N (clone 1 and clone 2) cells. Blots were stained for VPS35 and β-actin (total protein loading control)