Figure 3.
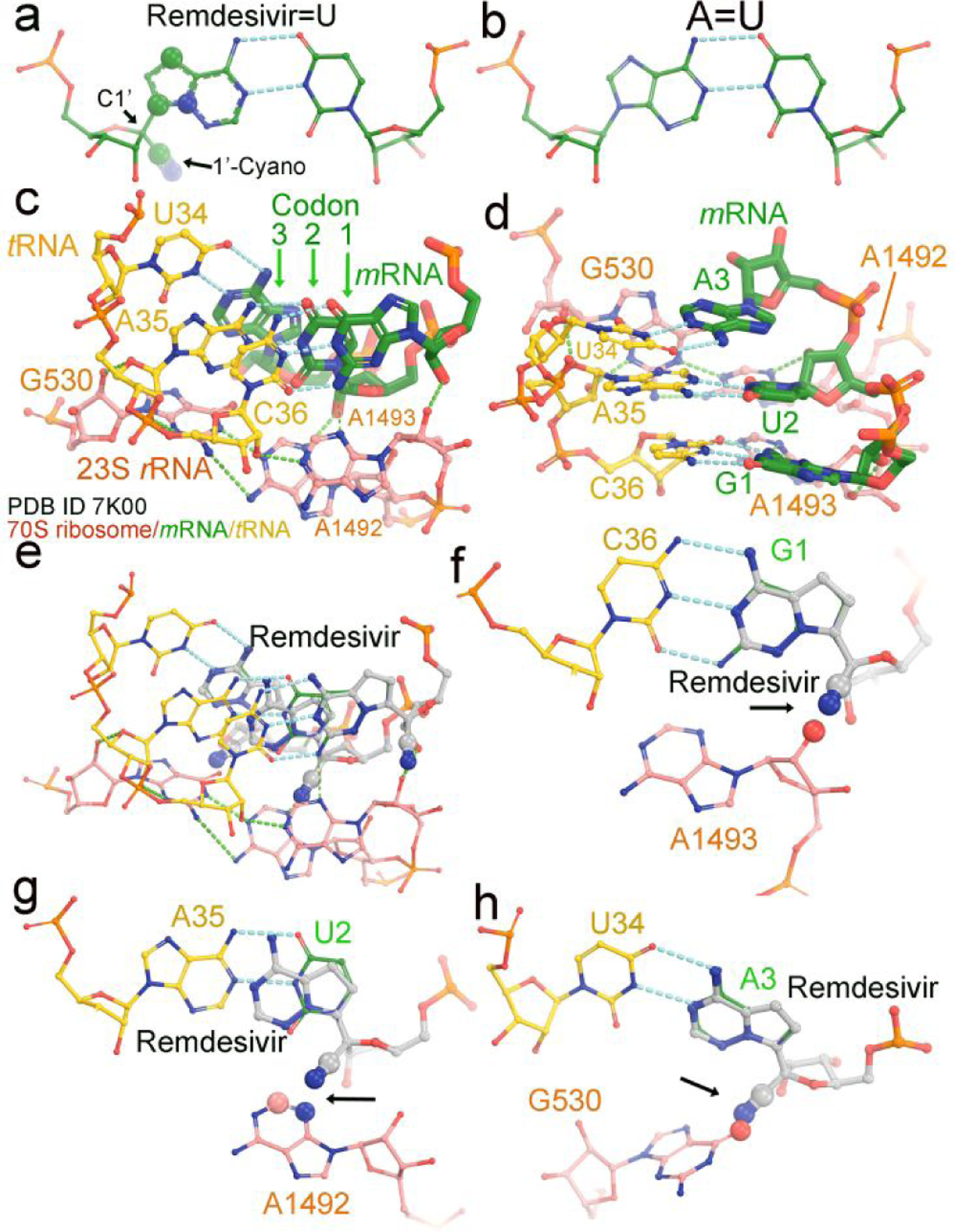
Modeling of remdesivir on a translating ribosome. (a) Base-pairing between RMP and UMP. The 1’-cyano substitution at C1’ and three other substitutions in the nucleobase are in large spheres (30% of van der Waals radii). (b) Watson-Crick AMP=UMP base pair. (c, d) Two views of the translating 70S E. coli ribosome cryo-EM structure (PDB ID 7k00) in complex with mRNA (green) and tRNA (yellow) at the decoding center. Three nucleotides of 23S rRNA, G530, A1492 and A1493 are shown in salmon. The three codon nucleotides in the structure are G1, U2 and A3 and in green. Three tRNA anticodon nucleotides are U34, A35 and G36 and in salmon. (e-h) Modeled remdesivir at the first, second, and third codon positions. Large spheres and arrows show where severe clashes occur.
