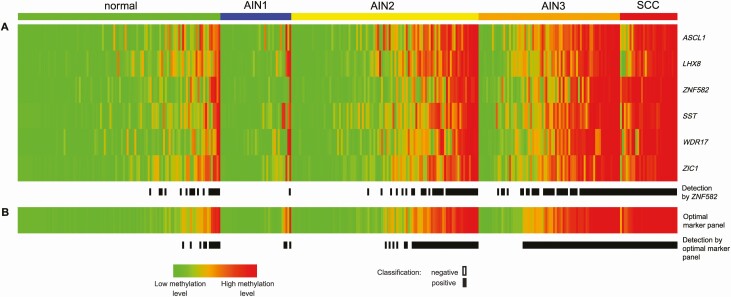
Figure 2.
A, DNA methylation pattern of the 6 individual methylation markers (rows: ASCL1, LHX8, SST, WDR17, ZIC1, ZNF582; based on LOOCV univariable regression for AIN3+ detection), per histological category in cross-sectional series. Methylation result per sample (column) in the different histological subgroups is displayed in color according to the PP from green (low methylation levels; PP of 0) to red (high methylation levels; PP of 1). In each group, samples are ordered based on their average predicted probability. Black boxes [detection by ZNF582] represent samples classified as methylation positive using ZNF582 alone (at the non-CV Youden index threshold ≥0.35). B, DNA methylation pattern of the optimal marker panel (ASCL1, SST, ZNF582; based on LOOCV multivariable regression for AIN3+ detection) and methylation positivity using the panel. Black boxes [detection by optimal marker panel] represent samples classified as methylation positive using the optimal marker panel (at the non-CV Youden index threshold ≥0.43). Endpoint: AIN3+ (AIN3 and anal SCC). Abbreviations: AIN1–AIN3, anal intraepithelial neoplasia (grades 1–3); LOOCV, leave-one-out cross-validated; non-CV, non–cross-validated; normal, normal control samples; PP, predicted probability; SCC, squamous cell carcinoma.