Figure 1.
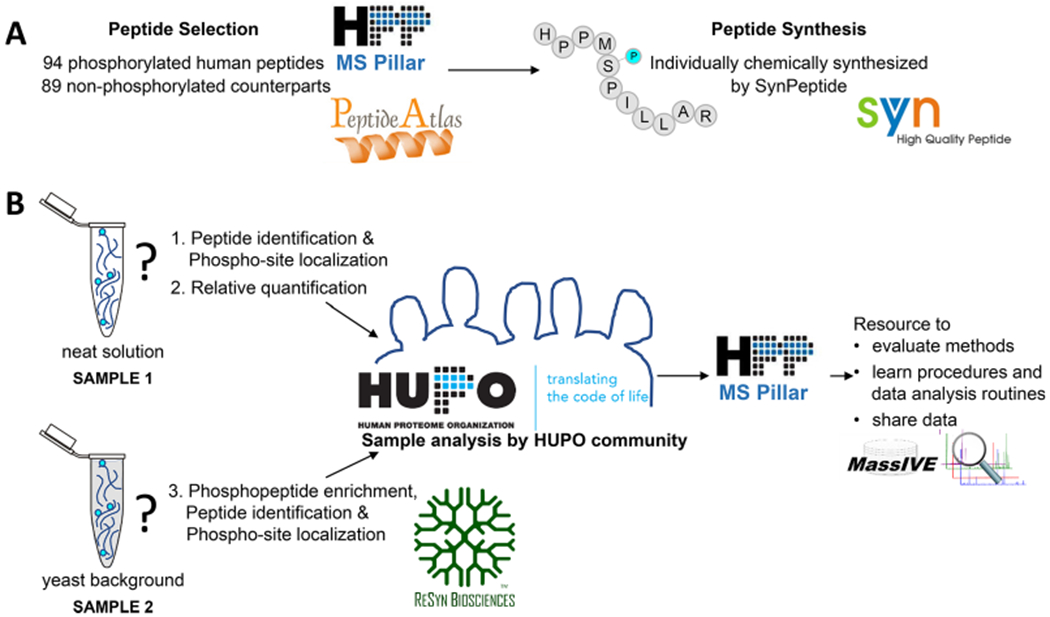
Overview of the HPP Phosphopeptide Challenge. (A) The MS Resource Pillar of the Human Proteome Project (HPP) selected 94 human phosphorylated peptides (number unknown to participants) and 89 nonphosphorylated counterparts (sequences provided to participants) considering highly observed peptides in PeptideAtlas and applied the criteria described in the Experimental Section and in Figure 2. Synthetic peptides were graciously provided by SynPeptide (www.synpeptide.com). (B) 94 phosphorylated and 89 nonphosphorylated peptides were mixed at different ratios and provided to the HUPO community as a neat solution and in a yeast background. Participants were asked to use their method(s) of choice to (1) identify the number and location of phosphorylation sites for all phosphorylated peptides in the mixture, (2) determine the relative abundance of each phosphopeptide compared with its nonphosphorylated counterpart in the neat sample, and (3) enrich for the phosphorylated peptides spiked into a yeast background and report the human phosphopeptides that were detected. ReSyn Biosciences (www.resynbio.com) graciously provided optional phosphopeptide enrichment kits to participants. The goal of the HPP MS Resource Pillar Phosphopeptide Challenge was to help the community evaluate methods, learn procedures, and improve data analysis routines for phosphopeptides. All data from this collaborative effort are shared through MassIVE and serve as resources for the development of new computational tools. SynPeptide logo reprinted with permission from SynPeptide Co., Ltd. HUPO and HPP MS Pillar logos reprinted with permission from the Human Proteome Organization. MassIVE logo reprinted with permission from Computer Science and Engineering, University of California, San Diego. ReSyn logo reprinted with permission from ReSyn Biosciences. PeptideAtlas logo reprinted with permission from the Institute for Systems Biology.
