Figure 1.
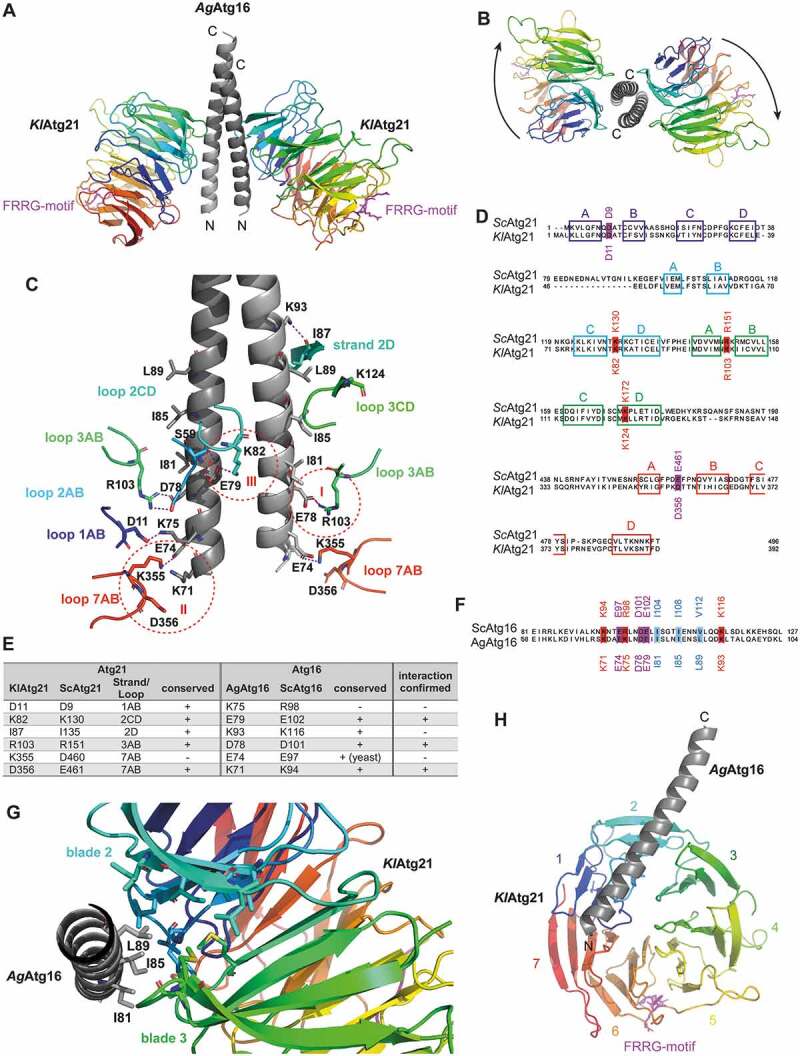
Structure of the Atg16-Atg21 complex. (A) Crystal structure of K. lactis Atg21 (rainbow-colored: blue to red from amino to carboxy terminus) in complex with the coiled-coil domain of A. gossypii Atg16(40–124) (light gray). The crystal structure was solved up to a resolution of 3.7 Å. The conserved lipid-binding motif FRRG of KlAtg21 is labeled in pink. (B) View from the top on the carboxy-terminus of the AgAtg16 coiled-coil domain and the KlAtg21 beta-propellers. Arrows indicate the orientation of the propeller blades. (C) Putative polar contacts between the AgAtg16 coiled-coil domain and KlAtg21 were predicted using PyMOL. Red dashed circles highlight three potential salt bridges (I–III). (D and F) Sequence alignment of Sc and KlAtg21 (D) or Sc and AgAtg16 (F), respectively, using Jalview and MAFFT (Clustal color code [76]). The conservation of the potentially interacting residues of Atg21 and Atg16 is shown. Colored boxes represent the respective strands of Atg21. (E) Summary of the potential polar contacts between AgAtg16 and KlAtg21, as well as their conservation and counterparts in S. cerevisiae. (G) View from the carboxy-terminus of the monomeric AgAtg16 alpha-helical domain. Residues I81, I85, and L89 of AgAtg16, which are located in three consecutive alpha-helical turns form a hydrophobic patch that inserts into a hydrophobic cleft of KlAtg21 formed between blades 2 and 3. (H) View on the bottom side of the KlAtg21 beta-propeller that shows the position of the monomeric Atg16 coiled-coil domain at the beta-propeller
