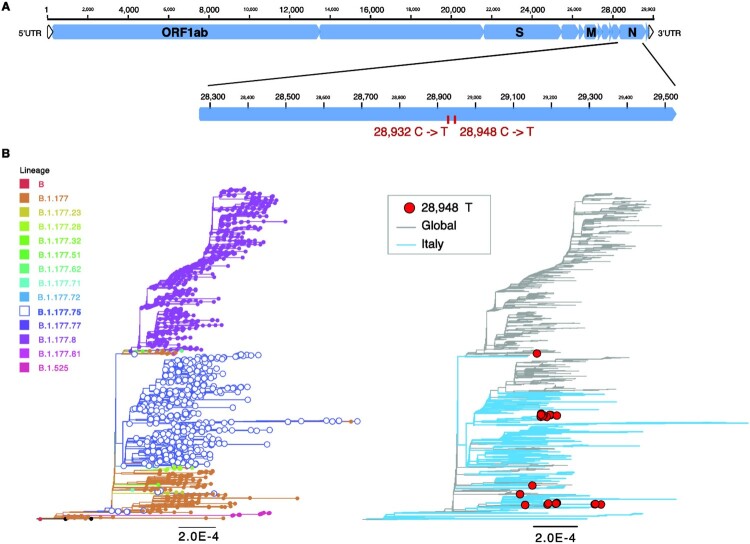
Figure 1.
Genome organization of SARS-CoV-2 and variant specific mutations. (A) Shown is a schematic view of the full genome of SARS-CoV-2 (top) and the N gene enlarged (bottom) to simplify visualization of the two mutations C to T nucleotide substitutions at position 28,932 and 28,948 (ref accession NC_045512). (B) Maximum likelihood (ML) trees for SARS-CoV-2 full genomes (n=1858) including lineages B.1.177 (n=246), B.1.177.8 (n=910), B.1.177.75 (n=642), and others (n=62). Italian sequences (n=567) are shown in blue and global sequences in grey. Lineage specific mutations are indicated. Sequences containing mutant variant for 28,948 in red (right). Branch length indicates nucleotide substitutions per site. Tree is rooted at the reference sequence NC_045512.