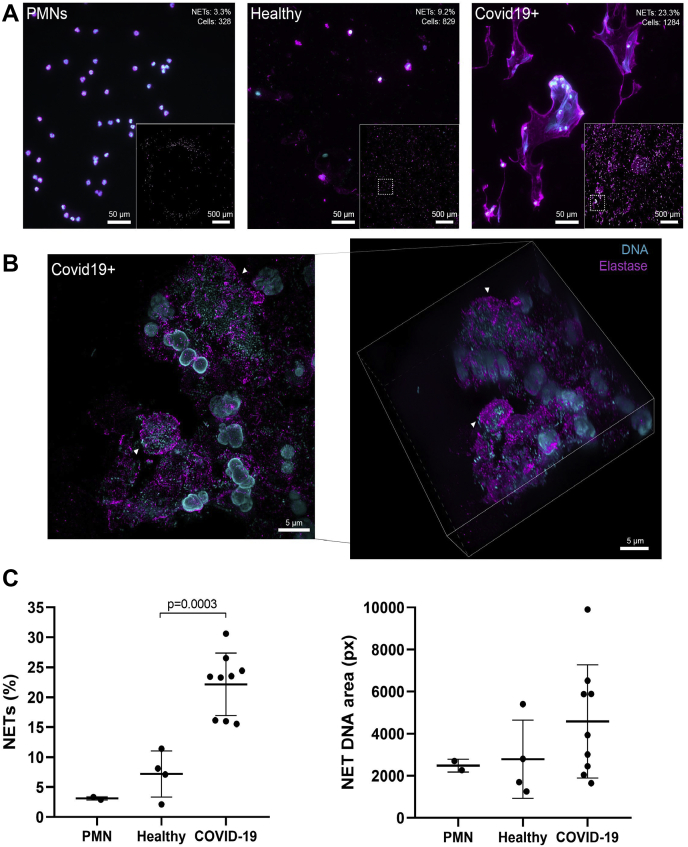
Fig. 2.
NETs in the sputum of SARS-CoV2 positive who were not treated with rhDNase and healthy controls.A, widefield microscopy of neutrophil elastase (magenta, antielastase antibody) and DNA (cyan, DAPI) in sputum samples or purified neutrophils. Insets show stitched overview image from 6 × 6 images of the samples. Quantification of single cell–level NETs (percent of cells and number of cells) of each overview is shown in the top corner of representative images. B, super-resolution microscopy of neutrophil elastase (magenta, antielastase antibody) and DNA (cyan, DRAQ5) in a nontreated patient sample. Left, maximum intensity projection of structured illumination microscopy Z-stack. Right, isometric view of Z-stack. Arrowheads show either a fully formed NET (top) or a neutrophil with decondensed DNA undergoing NETosis (bottom). C, quantitative data of A are shown as the percentage of NET-positive cells and the corresponding average area that the DNA signals covers in each cell. Each point indicates quantification of 36 images from independent samples. Lines indicate the mean for each category. DAPI, 4,6 diamidino-2-phenylindole; NETs, neutrophil extracellular traps; rhDNase, recombinant human DNase I; SARS-CoV2, severe acute respiratory syndrome coronavirus 2.