Fig. 2.
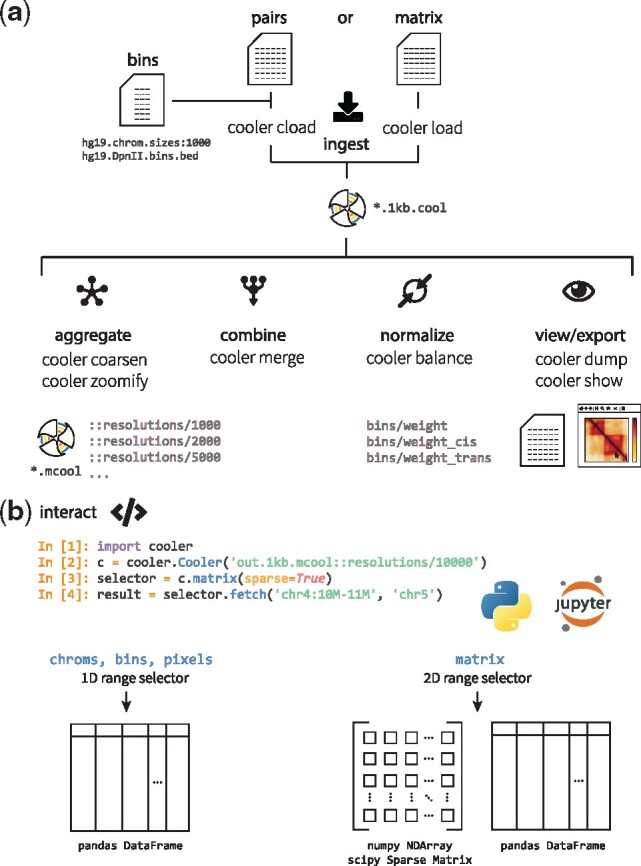
Cooler CLI and Python library. (a) Summary of the main categories of cooler commands available with the cooler Python package, illustrating the flow of data. The main operations include the ingestion of file or text streams to create new coolers, aggregation and coarsening of existing coolers to lower resolutions, merging of axis-compatible matrices, normalization of cooler matrices by iterative correction, utilities to serialize and stream out the data and metadata inside a cooler file and to process range queries and a lightweight viewer to visually inspect a matrix. For example, one uses either the load command to ingest pre-aggregated data already in matrix form or the cload command to aggregate paired tag records into a matrix. The genomic bin segmentation defining the axes of the matrix must be provided separately by providing either a path to a BED file or a path to a chromosome sizes file along with a specified fixed bin size. (b) The cooler Python library provides a Cooler class that exposes data range selectors to facilitate data retrieval and analysis. The individual chrom, bin and pixel tables are accessible using 1D range selectors that accept column and row-range selections and yield pandas data frame output. A cooler’s matrix values are also exposed using a 2D range selector that processes rectangular range queries specified either by a pair of genomic coordinate intervals in UCSC-style notation (using the fetch method) or as integer matrix coordinates (using Python slice syntax). The retrieved 2D range data may be materialized as dense NumPy arrays, sparse matrices or data frames. For symmetric coolers, the file’s upper triangular data will be appropriately mirrored in the array and sparse matrix outputs
