Figure 2.
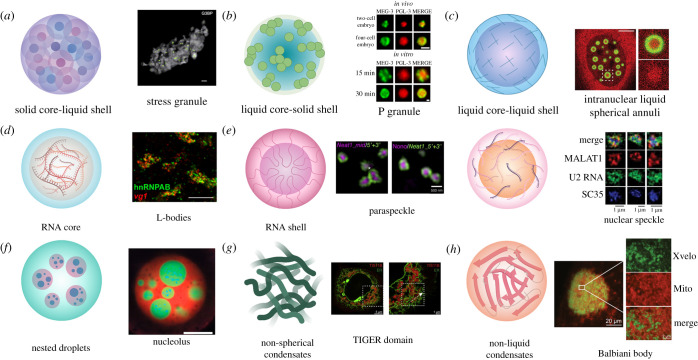
Examples of biomolecular condensate architecture. (a) Solid core-liquid shell. Condensates with this structure initially form as individual protein cores, which then accumulate more material and join together to form larger structures, as with SGs. Green dots indicate G3BP cores and grey represents the surface area of the SG; scale bar represents 500 nm [83]. (b) Liquid core-solid shell. Condensates with this structure consist of a more liquid-like core surrounded by a gel-like protein shell. An example of this architecture is the P granule found in C. elegans embryos. Condensates formed by MEG-3 (green) and PGL-3 (red) in cells and in vitro show this architecture; scale bars represent 1 µm [84]. (c) Liquid core-liquid shell. With this architecture, the interior and the exterior of the condensate are both dynamic liquids, but are compositionally distinct, such as with iLSA. iLSA have a liquid TDP-43 shell (green) and the interior is enriched for HSP70 chaperones, including Hsc70 (red); scale bar represents 5 µm [85]. (d) RNA core. In X. laevis oocytes, RNAs must properly localize for successful embryonic development. This localization is achieved, in part, by the formation of L-bodies, composed of RNAs and RBPs. In L-bodies, the RNA forms an immobile core enveloped by a dynamic protein coating. Staining for the L-body protein hnRNPAB (green) and mRNA vg1 (red) reveals the multiphasic nature of these condensates; scale bar represents 10 µm [86]. (e) RNA shell. There are multiple ways in which a condensate can form such that protein is surrounded by RNA. In paraspeckles (left), the lncRNA NEAT1 folds so that its 3′ and 5′ ends face outward with its middle at the centre where proteins accumulate. Labelling the 5′ and 3′ ends of NEAT1 (green) highlights the paraspeckle shell, whereas labelling the middle portion of NEAT1 or a protein such as Nono (purple) identifies the core; scale bar represents 500 nm [87]. Alternatively, RNAs can decorate the exterior of a protein core, as with nuclear speckles (right). In this situation, a proteinaceous core collects RNA molecules that reside on the outside of the condensate. SIM images of nuclear speckles show splicing protein SC35 (blue) occupying a central area, with lncRNA MALAT1 (red) and U2 RNA (green) taking up an area with a larger radius; scale bars represent 1 µm [33]. (f) Nested Droplets. The nucleolus is an example of a condensate with a nested droplet architecture. The nucleolus has a tripartite organization, with the gel-like FC and DFC enveloped by a liquid-like GC. Each component of the nucleolus has a distinct function and displays distinct physical properties. In X. laevis nuclei, RNA Pol I (blue) localizes to the innermost FC, FIB1 (green) stains the DFC, and NPM1 (red) is localized at the liquid-like GC; scale bar represents 20 µm [88]. (g) Non-spherical condensates. Biomolecular condensates can also adopt atypical structures, as in the case of TIS11B granules. TIS11B granules form an extensive mesh-like assembly composed of protein and RNA. In cells, TIS11B condensates (red) intertwine with the ER (green), forming a complex network of tubules; scale bars indicate 5 µm (left) and 1 µm (right) [89]. (h) Non-liquid condensates. In addition to liquid-like condensates, biomolecules can also condense to form solid states. One example of this is the Balbiani body, which contains an amyloidogenic network formed by the protein Xvelo in X. laevis (homologous to the Bucky ball protein in zebrafish). Xvelo (green) contains a PrLD which is necessary for its self-assembly, as well as its association with organelles, like mitochondria (red); scale bars indicate 20 µm and 2 µm (insets) [90]. This figure was made with BioRender.