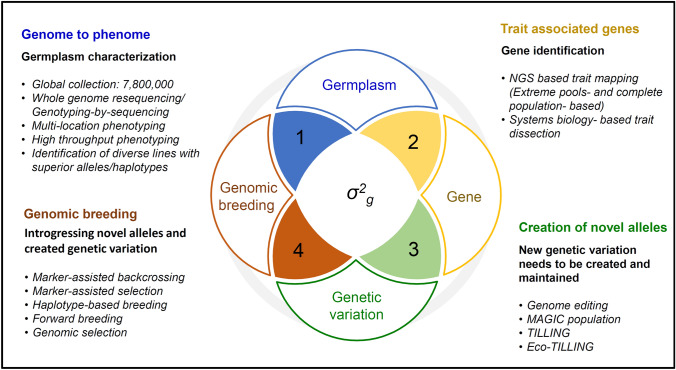
Fig. 1.
Enhancing genetic variation through identifying/ creating and utilizing favorable alleles/haplotypes. Genetic variance (σ2g) is estimated with genetic parameters including phenotypic variance composed by genotypic and environmental variance (not inheritable). Genetic variance allows an understanding of the genetic structure involved in the progenies, determined by additive and non-additive effects. (1) Genome to phenome deals with the connection and causation between the genetic makeup of an accession and the observed physical or physiological traits or characteristics (phenome). This can be achieved by characterizing germplasm collections at the phenotypic and genotypic level. (2) Trait associated genes can be identified through NGS based trait mapping (extreme pools- based and complete population-based) and/or through systems biology approaches. (3) Genetic variations can be assayed using germplasm characterization or can be created by through genome editing, multiparent advanced generation intercross (MAGIC), targeting induced local lesions in genomes (TILLING) and Eco-TILLING populations. (4) Genomic breeding to combine superior/ novel alleles/ haplotypes in elite backgrounds. The identified genetic variations can be introduced in a crop improvement programs through genomics-assisted breeding (GAB) approaches including marker- assisted backcrossing (MABC), marker- assisted recurrent selection (MARS), haplotype-based breeding (HBB), forward breeding (FB) and genomic selection (GS).