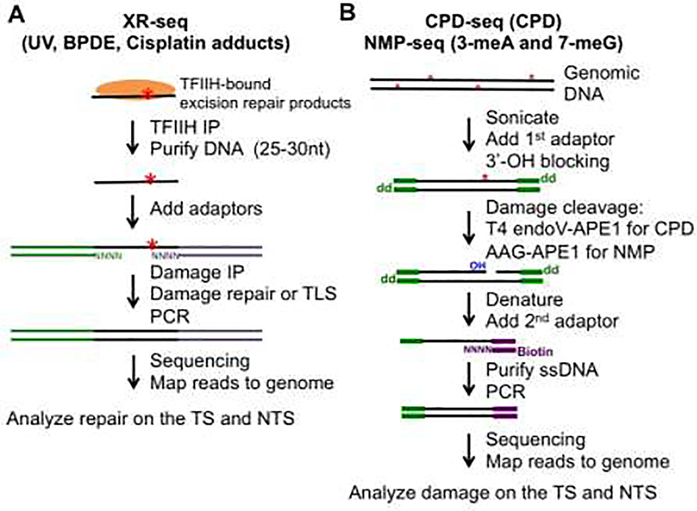
Figure 2.
Schematic representation of DNA damage/repair mapping methods. (A) XR-seq is a method that maps excision repair products (~30 nt), which are bound by TFIIH in the cell. These short fragments are extracted from cell lysates and ligated to sequencing adaptors. Fragments containing damage are purified by damage immuneprecipitation (IP) using a DNA damage-specific antibody. After damage repair or bypass with a translesion synthesis (TLS) DNA polymerase, the DNA fragment is used for PCR amplification. (B) CPD-seq and NMP-seq are used for mapping UV damage (i.e., CPDs) and alkylation lesions such as 3-meA and 7-meG. Red asterisks represent DNA damage. Damage is cleaved with DNA repair enzymes to generate a new 3’-OH group, which is ligated to a sequencing DNA adaptor (2nd adaptor).