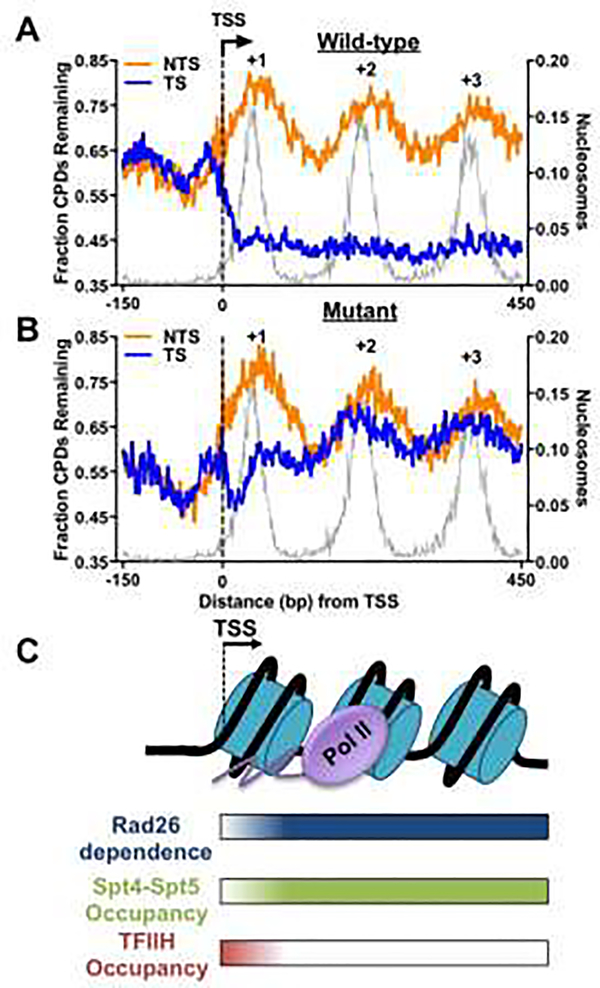
Figure 3.
Role of Rad26 in yeast TC-NER revealed by CPD-seq analysis. (A) Repair of CPDs in wild-type cells at 2h. Yeast genes (n = 5,205) [74] were aligned at their transcription-start site (TSS) and the fraction of remaining CPDs at 2h (CPD-2h normalized to CPDs-0h) was plotted in DNA regions around the TSS. Transcribed strand (TS) and non-transcribed strand (NTS) are analyzed separately. CPD-seq data was downloaded from Gene Expression Omnibus (accession code GSE145911) [59] and reanalyzed in transcribed regions near the TSS (from −150bp upstream to 450bp downstream of the TSS). The gray background depicts yeast nucleosome occupancy generated with published MNase-seq data [75]. (B) Repair of CPDs in Rad26-deficient cells (i.e., rad26Δ) at 2h in yeast genes. (C) Model depicting the variable requirements for Rad26 in yeast TC-NER in different chromatin regions (e.g., +1 and downstream nucleosomes), and its correlation with the occupancy of transcription elongation factor Spt4-Spt5 and initiation/repair factor TFIIH.