Figure 1.
Copy number variation at thousands of VNTRs is associated with variation in gene expression and DNA methylation in cis
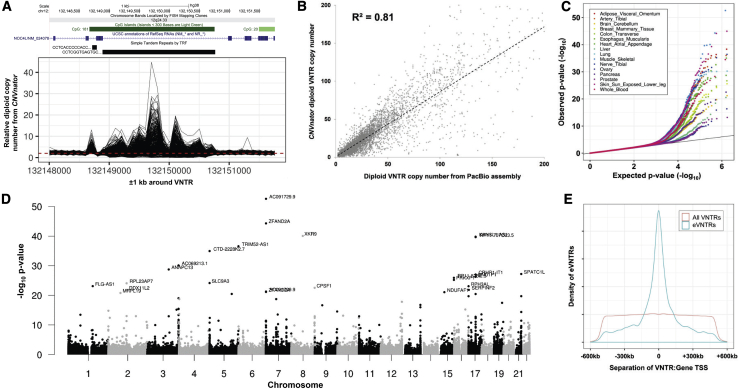
(A) CNVnator-estimated copy number per 100 bp bin over a VNTR region shows highly variable read depth among samples from the GTEx cohort. Shown is read depth data for a 44-mer repeat that has 43 copies in the reference genome (chr12: 132,148,891–132,150,764, hg38), located intronic within NOC4L, which shows >10-fold difference in copy number within the population.
(B) Read depth provides good accuracy for estimating diploid VNTR copy number. Using 14 samples where both Illumina and PacBio WGS data were available, at 1,891 eVNTR loci we compared diploid VNTR copy number estimates from WGS read depth by using CNVnator with direct genotypes derived from Pacific Biosciences long-read diploid assemblies. We observed a high correlation between the two approaches (R2 = 0.81).
(C) Q-Q plots showing the distribution of observed versus expected p values for eVNTRs in 16 representative GTEx tissues. Variations in the observed p value distribution among GTEx tissues are a reflection of the varying sample sizes available, which strongly influence statistical power.
(D) Manhattan plot showing results of cis-association analysis between VNTR copy number and gene expression in skeletal muscle samples from the GTEx cohort. The high frequency of significant associations in subtelomeric and centromeric regions is consistent with the known enrichment of VNTRs in these regions.30,31
(E) Significant eVNTRs are highly enriched within close proximity to the genes whose expression level they are associated with, mirroring similar observations made for SNV eQTLs.32,33 We also observed a similar relationship for mVNTRs and the CpGs they associate with (Figure S10), although we note an approximate order of magnitude difference in the distances over which significant eVNTRs and mVNTRs were typically observed to function.