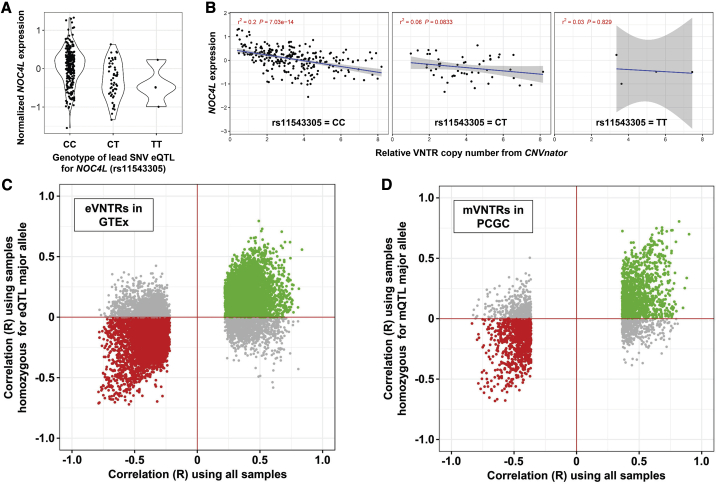
Figure 3.
Copy number variation at the majority of VNTRs shows association with gene expression and DNA methylation independently of SNV eQTLs and mQTLs
We performed conditional analysis of eVNTRs and mVNTRs after removing the effect of the strongest SNV QTL on the same target. Shown is an example locus, a 44-mer repeat that has 43 copies in the reference genome (chr12: 132,148,891–132,150,764, hg38), corresponding to the same VNTR shown in Figure 1A. This VNTR is located intronic within NOC4L and is significantly associated with NOC4L expression.
(A) We identified rs11543305, a C/T variant that is located 1.6 kb proximal to the VNTR, as being the lead SNV associated with NOC4L expression.
(B–D) After stratifying samples on the basis of genotype at rs11543305, copy number of this VNTR still shows a significant association with NOC4L expression (B). Considering all significant VNTRs we identified, including eVNTRs observed in GTEx (C) and mVNTRs observed in the PCGC cohort (D), there is a clear trend where the majority of observed VNTR associations retain their original signal even after conditioning on the genotype of the lead SNV QTL. These data indicate that the majority of VNTR associations we identified act independently of local SNV QTLs. In each plot, colored points represent VNTR associations that retain the same directionality after conditioning on the lead SNV QTL: either positive associations (green) or negative associations (red).