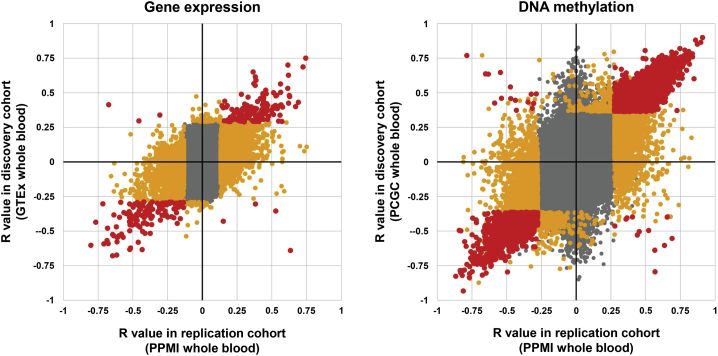
Figure 4.
Replication of the majority of significant eVNTRs and mVNTRs in an independent cohort
We performed replication analysis in the PPMI cohort, which comprises 712 individuals, with Illumina WGS, DNA methylation, and RNA-seq data derived from whole blood. We observed that 73% of significant eVNTRs detected in GTEx whole blood were also identified as significant in the PPMI cohort. Similarly, 80% of significant mVNTRs detected in the PCGC discovery cohort were also significant in the PPMI cohort. Points shown in gray were non-significant in both discovery and replication cohorts, points in orange were significant in one cohort, while points in red were significant in both cohorts.