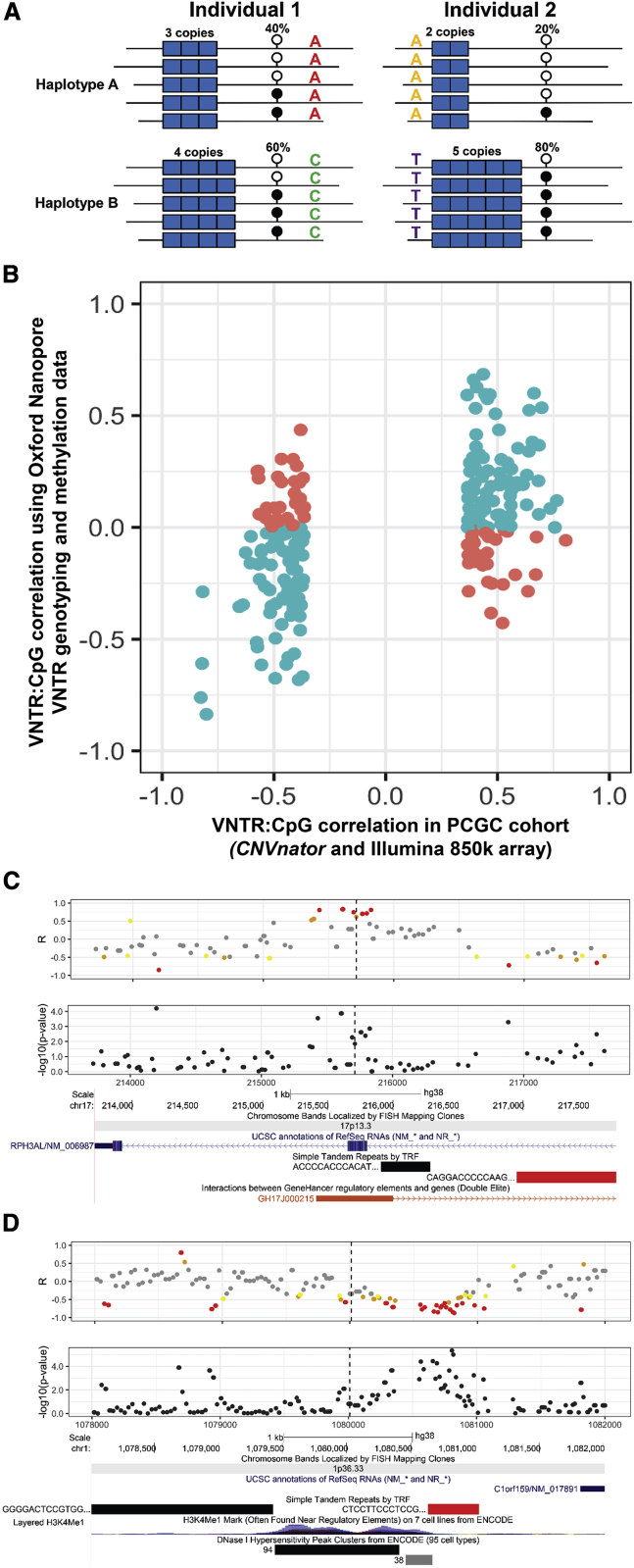
Figure 5.
Additional replication of mVNTRs from direct VNTR genotyping and methylation profiling in 30 genomes sequenced with Oxford Nanopore long reads
(A) Outline of how phased long reads can be used to perform allelic association analysis of VNTR genotype with cis-linked CpG methylation levels. In each individual, ONT reads are phased into the two haplotypes via SNVs (colored letters), VNTRs (blue blocks) are genotyped directly on each haplotype based on the phased assemblies, and CpG methylation levels (lollipops) on each haplotype are estimated on the basis of electrical current signals from each phased read.
(B) For mVNTR:CpG pairs identified in the PCGC discovery cohort that had ≥20 haplotypes each with ≥10× coverage in the 30 available ONT genomes, 163 of 228 (71%) showed the same directionality of association in this independent dataset.
(C) Copy number of an 83-mer VNTR (chr17: 216,953–218,561, hg38, indicated by the red bar) that lies intronic within RPH3AL is positively associated with local DNA methylation, including an annotated enhancer of RPH3AL. This same VNTR was negatively associated with RPH3AL expression in 22 GTEx tissues.
(D) Copy number of a 32-mer VNTR (chr1: 1,080,637–1,081,029, hg38, indicated by the red bar) that lies ∼800 bp upstream of C1orf159 is negatively associated with local DNA methylation, including a region of H3K4 mono-methylation and DNaseI hypersensitivity. This same VNTR was positively associated with C1orf159 expression in six GTEx tissues. In (C) and (D), plots show the correlation (R) values and unadjusted p values between VNTR copy number and CpG methylation measured directly from ONT reads. The dashed vertical lines indicate the position of a CpG that was associated with VNTR copy number in the PCGC discovery cohort. Correlation values are colored according to their significance in the 30 ONT genomes: yellow indicates p < 0.1, orange p < 0.05, and red p < 0.01. Below the plots are screenshots from the UCSC Genome Browser showing annotations of RefSeq genes, simple repeats, and regulatory regions.