Figure 1.
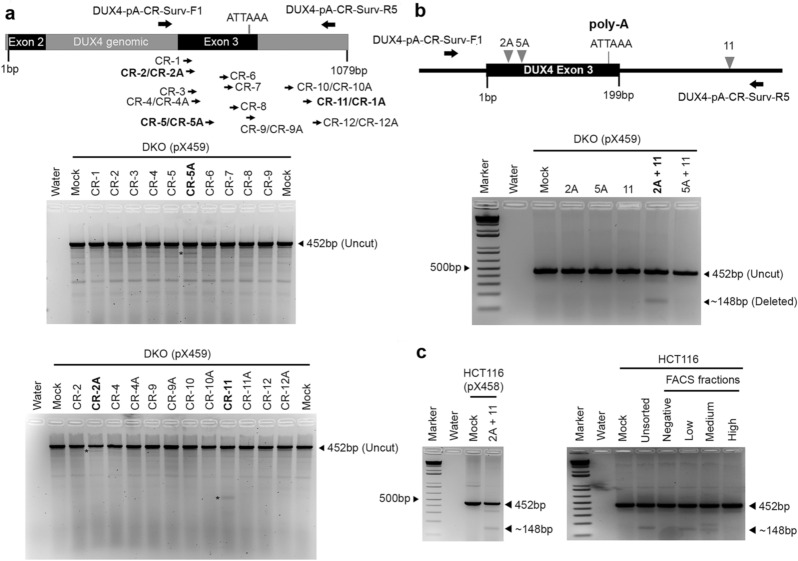
Identification of suitable sgRNAs to target DUX4 poly-A signal in HCT116 and DKO. (a) Top panel: Schematic of binding sites for sgRNAs (CR-1 to CR-12) across DUX4 exon 3 and a region downstream of it, which flank and span the poly-A site. sgRNAs with suffix ‘A’ are truncated (17–18 bp) versions of regular sgRNAs (20 bp) with same numerical value. The position of primers used for Surveyor detection of cut and uncut products are shown as black arrows, along with their names. The position of the pathogenic poly-A signal is also indicated. Arrows for efficient sgRNAs are highlighted in bold. The base pair length is indicated to provide a scale. Bottom panels: Ethidium bromide stained agarose gels depicting results of Surveyor assay for these sgRNAs. Sample names are indicated on top along with cell line used for transfection and vector in which sgRNAs were cloned. The expected size for uncut PCR product is indicated on the right-hand side by a black triangle. Efficient sgRNAs are highlighted in bold and the bands representative of their cutting is indicated by an asterisk on the left of the lane corresponding to a given sgRNA (b) Top panel shows schematic of trial for a dual-sgRNA PCR based deletion assay with suitable combinations sgRNAs upstream (CR-2A or CR-5A) and downstream (CR-11) of the poly-A site. The position of primers used for PCR detection of cut and uncut products are shown as black arrows, along with their names. Bottom panel shows results of deletion assay using abovementioned sgRNAs. Sample names are indicated on top along with cell line used for transfection and vector in which sgRNAs were cloned. The expected size for undeleted and deleted PCR products is indicated on the right-hand side by black triangles. The most efficient sgRNA combination is highlighted in bold. (c) Left panel shows results of a trial deletion assay using abovementioned sgRNAs cloned in pX458 and transfected in parental HCT116 cells. Bottom right panel shows results of deletion assay using abovementioned sgRNAs cloned in pX458 in HCT116 cells harvested before (Unsorted) or after (Negative, Low, Medium and High fluorescence intensity above background) FACS based sorting for GFP-positive cells. Sample names are indicated on top. Schematic diagrams were generated using Microsoft PowerPoint 16.48 (https://www.microsoft.com) and Adobe Photoshop 21.0.1 (https://www.adobe.com).