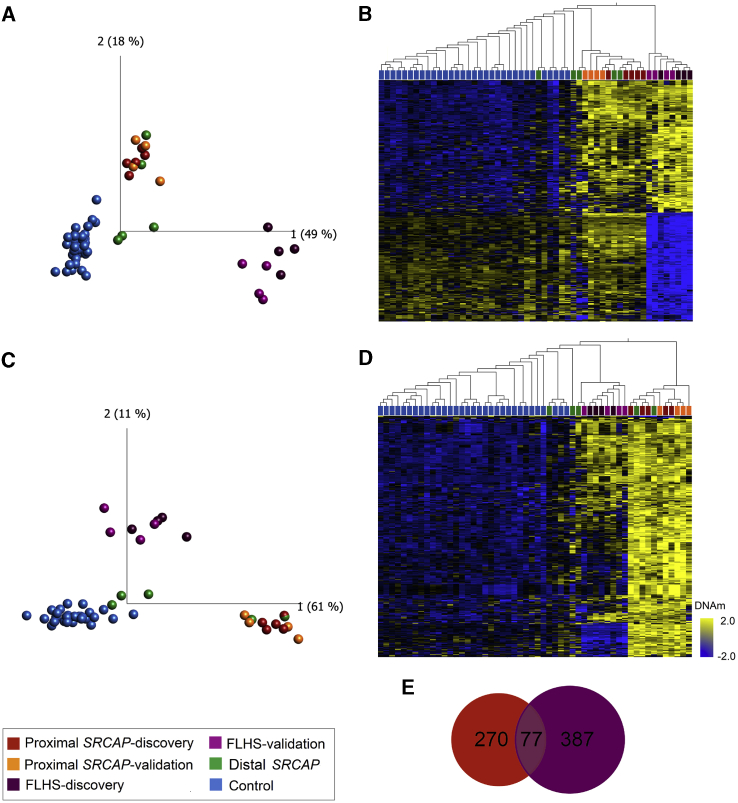
Figure 2.
Loss-of-function variants in SRCAP are associated with two distinct but overlapping DNAm signatures based on variant position
(A and B) FLHS DNAm signature: (A) principal components analysis (PCA) and (B) heatmap showing clustering of FLHS discovery subjects (n = 4; dark purple), FLHS validation subjects (n = 4; light purple), proximal SRCAP discovery subjects (n = 5; dark orange), proximal SRCAP validation subjects (n = 4; light orange), distal SRCAP subjects (green) and discovery control subjects (n = 35; blue) using DNAm values at 464 CpG sites in the FLHS DNAm signatures. FLHS subjects clearly segregate from all other samples, while all proximal and some distal SRCAP cases cluster intermediately.
(C and D) Proximal SRCAP DNAm signature: (C) PCA and (D) heatmap showing clustering of the same subjects from (A) and (B) and matched control subjects (n = 32; blue) using DNAm values at 347 CpG sites in the proximal SRCAP DNAm signature. Proximal SRCAP discovery and validation subjects and some distal subjects clearly separate from control subjects, with FLHS subjects clustering intermediately. The heatmap color gradient indicates the normalized DNAm value ranging from −2.0 (blue) to 2.0 (yellow). Euclidean distance metric is used in the heatmap clustering dendrograms.
(E) Venn diagram showing the CpG sites are shared and distinct between the proximal SRCAP and FLHS signatures.