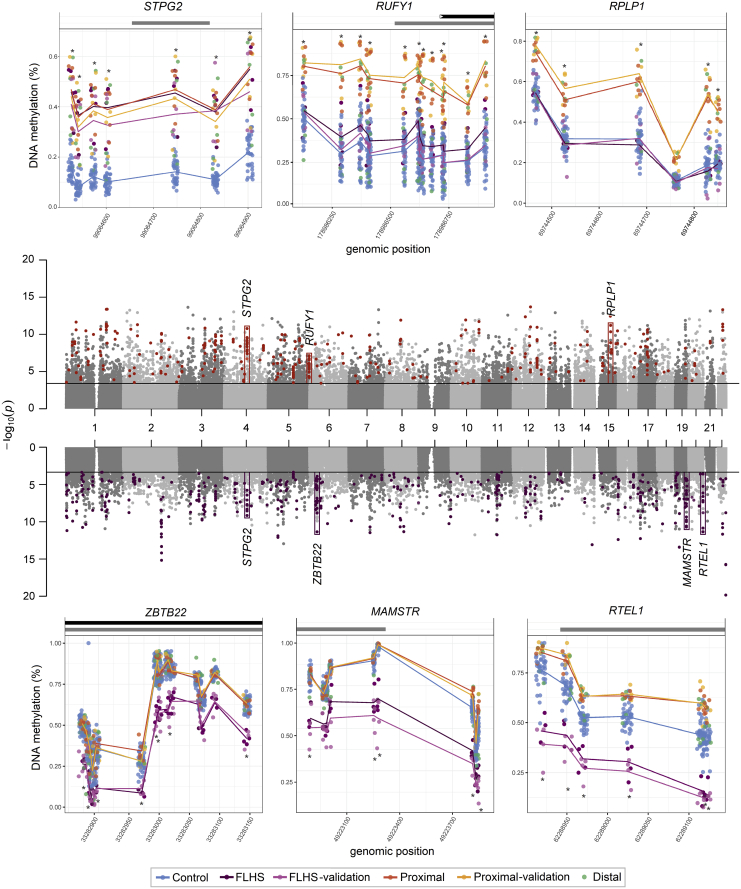
Figure 3.
Distribution of differentially methylated regions in SRCAP DNAm signatures
The Manhattan plots (center) show the CpG site p values for each DNAm signature discovery comparison; sites that meet the |Δβ| > 0.20 cut off in the proximal SRCAP signature (upper) are colored orange, those that meet the cut off in the FLHS signature (lower) are colored purple. Six differentially methylated regions are boxed on the Manhattan, with the full plots for sample groups shown above and below. These six illustrate different patterns of DNAm between the proximal SRCAP and FLHS groups. Each is named for the gene body/promoter to which they map. Lines connect the average beta values for each corresponding group. Grey bars above the plots indicate CpG islands, black represent gene bodies. β values for each sample are indicated with mean lines for groups (except the distal group, since two classified positively and three negatively). The CpGs mapping to STGP2 are present in both signatures, RUFY1 and RPLP1 are in the proximal SRCAP signature, and ZBTB2, MAMSTR, and RTEL1 are in the FLHS signature. There is increased DNAm at the RTEL1 sites in the Proximal group, though not a large enough Δβ to be in the proximal SRCAP DNAm signature. In all cases, β values in the discovery and validation cohorts are concordant. RPLP1, RUFY1, and RTEL1 are encoded on the plus strand, STPG2, ZBTB22, and MAMSTR are on the minus strand.