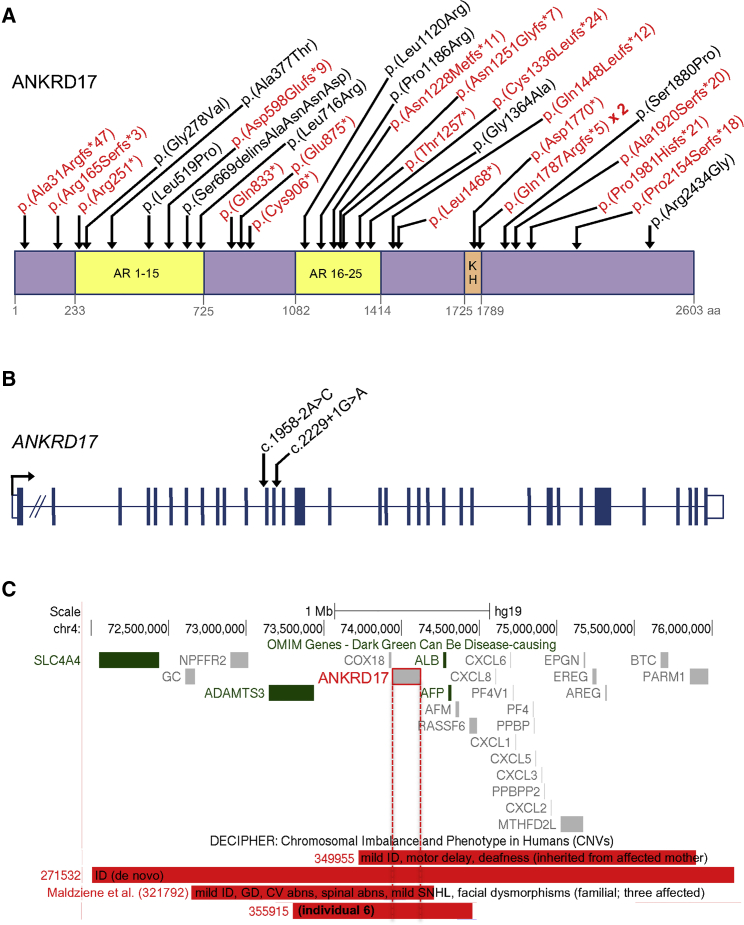
Figure 1.
Distribution of pathogenic ANKRD17 variants and 4q13.3 deletions
(A) Variants affecting coding sequence. Truncating variants are in red. Domain boundaries are based on Uniprot entry O75179. AR, ankyrin repeats; KH, K Homology domain.
(B) Variants affecting essential splice sites. Exon-intron structure drawn approximately to scale (apart from intron 1), according to GenBank: NM_032217.4.
(C) Deletions of the region containing ANKRD17 on chromosome 4q13.3. Among the disease-associated OMIM genes in the interval (those in green), the mode of inheritance or associated phenotype is not compatible with that of the deletions shown. Only deletions under 5 Mb from the DECIPHER database are shown. For DECIPHER individuals 349955 and 271532, publicly available clinical information is listed; individual 321792 is one of three affected individuals of the previously described familial case10 (the phenotypes listed on the figure are a summary of all three family members); and individual 355915 corresponds to individual 6 in the present report (see Table S1 for details). ID, intellectual disability; GD, growth delay; CV, cardiovascular; abns, abnormalities; SNHL, sensorineural hearing loss.