Figure 4.
Modeling of sin3b disruption in zebrafish shows abnormal craniofacial patterning, reduced body length, and pathogenicity of case-associated missense variants
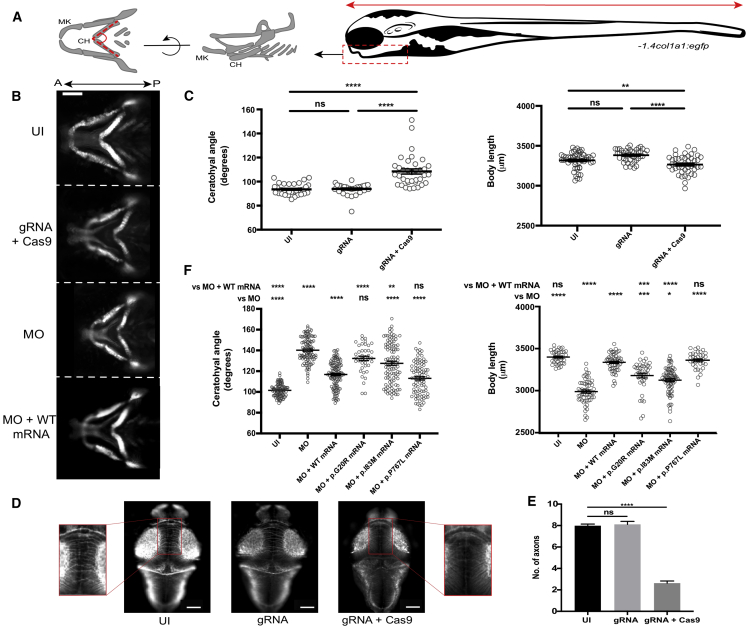
(A) Schematic of structures of interest used for in vivo measurements in developing zebrafish larvae at 3 days post fertilization (dpf); cartilaginous craniofacial structures (left, angle between dashed lines); body length (right, red horizontal arrow); CH, ceratohyal cartilage; MK, Meckel cartilage. −1.4col1a1:egfp transgenic zebrafish larvae were positioned with the Vertebrate Automated Screening Technology (VAST) BioImager and ventral fluorescent images were captured as indicated (right panel, red vertical arrow).
(B) Representative ventral images of in vivo models of sin3b disruption in zebrafish. CH angle was measured as indicated in (A). UI, uninjected; MO, morpholino; WT, human SIN3B mRNA. Scale bar, 100 μm.
(C) Quantification of craniofacial defects and body length in CRISPR/Cas9 F0 mutants. Small insertions and deletions were introduced into sin3b exon 13 via a high-efficiency guide RNA (gRNA). Targeting with gRNA leads to an increase of CH angle (left) and decrease in body length (right), indicating a crucial role for the Sin3 complex in the patterning of anterior structures and growth (n = 21–34 and 21–27 larvae/batch, repeated, for craniofacial and body length, respectively).
(D) Representative images showing fluorescent signal detected from the dorsal aspect of fixed and acetylated tubulin antibody-stained 3dpf larvae. Zoomed insets (left and right) show the region assessed for commissural neurons that cross the midline.
(E) Quantification of intertectal neurons in sin3b F0 mutants as a proxy for corpus callosum defects in SIN3B mutation-bearing humans. n = 49–50 larvae/batch, repeated.
(F) Transient suppression models mimic sin3b CRISPR F0 mutants. sin3b morphants (injected with 9 ng of e2i2 splice-blocking [sb] MO) display a broadened CH angle (left) and shortened body length (right) compared with uninjected controls. Coinjection of sin3b e2i2 splice-blocking MO with 100 pg of SIN3B WT human mRNA rescues this phenotype significantly. Coinjection of MO with case-specific variant mRNA (encoding p.Gly20Arg and p.Ile83Met) resulted in significantly reduced ability to rescue CH and body length compared with SIN3B WT mRNA (n = 41–61 larvae/batch, repeated). p.Pro767Leu is a negative control variant (rs117307745; six homozygotes in gnomAD). Statistical analyses were performed via a non-parametric Kruskal-Wallis test. ∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; ns, not significant; error bars represent standard error of the mean.