Figure 1.
AFF3 and AFF4 degron motif variants
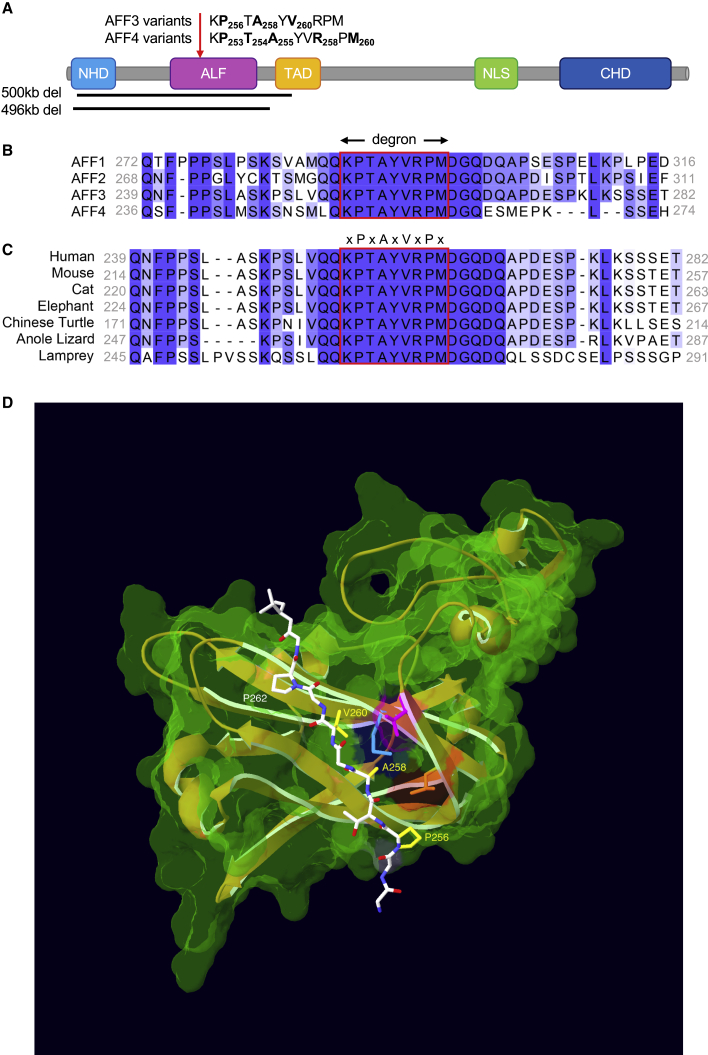
(A) Schematic protein structure of ALF proteins from the amino terminus: an N-terminal homology domain (NHD), the AF4/LAF4/FMR2 (ALF) homology domain23 containing the SIAH-binding degron motif, a serine-rich transactivation domain (TAD),3 a bipartite nuclear/nucleolar localization sequence (NLS), and a C-terminal homology domain (CHD). The sequences of the degron motifs of AFF3 and AFF4 are shown above. The residues modified in the KINSSHIP probands described in this manuscript and individuals affected by CHOPS18,19 are highlighted in bold and numbered. The extent of the 496 kb deletion identified in this work and the extent of the 500 kb deletion previously described38 are indicated by black bars. A red arrow pinpoints the position of the degron motif.
(B) Amino acid sequence alignment of human AFF1, AFF2, AFF3, and AFF4 proteins (ENSP00000305689, ENSP00000359489, ENSP00000317421, and ENSP00000265343, respectively) showing the highly conserved degron motif (red rectangle) of the ALF homology domain that provides the binding moiety to the SIAH ubiquitin-ligase. Sequence alignment was performed with Clustal Omega and edited with Jalview. Shading is proportional to conservation among sequences.
(C) Amino acid sequence alignment of different AFF3 vertebrate orthologs showing the conservation of the degron motif (red rectangle). Accession numbers are ENSP00000317421 (human), ENSMUSP00000092637 (mouse), ENSFCAP00000024603 (cat), ENSLAFP00000010776 (elephant), ENSPSIP00000007060 (chinese turtle), ENSACAP00000008035 (anole lizard), and ENSPMAP00000008605 (lamprey).
(D) 3D modeling of the binding of human AFF3 degron to the mouse Siah ubiquitin ligase. We downloaded PDB: 2AN6,35 in Swiss-PdbViewer34 and used it as a template to align the human SIAH ubiquitin ligase (UniProt: Q8IUQ4).33 With respect to the mouse crystal structure, the only difference is the presence of an aspartic acid residue instead of a glutamic acid at position 116. We then aligned the region of AFF3 containing the degron motif (LRPVAMVRPTV) onto the Siah-interacting protein46 peptide present in the crystal structure (QKPTAYVRPMD) to highlight the position of the variants reported in this study. For clarity, only sidechains of the core degron motif (Pro256, Ala258, Val260, and Pro262) are shown. The sidechains of KINSSHIP mutated residues are highlighted in yellow. The core degron motif adopts a beta-strand conformation directly contacting the ubiquitin ligase-binding groove. The sidechains of Ala258 and Val260 are embedded into binding pockets too small to accommodate larger sidechains.35 They are in direct proximity of Siah residues Thr156 (pink) and Met180 (cyan), identified as key binding residues in a series of pull-down assays.35 Replacing Pro256 with Leucine, a residue with a longer sidechain that will be positioned in proximity of Siah residue Leu158 (orange), could affect the backbone kink normally conferred by the conserved Proline. The longer sidechains of p.Pro256Leu, p.Ala258Thr, p.Ala258Ser, and p.Ala258Val variants and the smaller p.Pro256Ala and p.Val260Gly are likely to weaken or prevent the interaction with the ligase.