Summary
Clinical genetic testing of protein-coding regions identifies a likely causative variant in only around half of developmental disorder (DD) cases. The contribution of regulatory variation in non-coding regions to rare disease, including DD, remains very poorly understood. We screened 9,858 probands from the Deciphering Developmental Disorders (DDD) study for de novo mutations in the 5′ untranslated regions (5′ UTRs) of genes within which variants have previously been shown to cause DD through a dominant haploinsufficient mechanism. We identified four single-nucleotide variants and two copy-number variants upstream of MEF2C in a total of ten individual probands. We developed multiple bespoke and orthogonal experimental approaches to demonstrate that these variants cause DD through three distinct loss-of-function mechanisms, disrupting transcription, translation, and/or protein function. These non-coding region variants represent 23% of likely diagnoses identified in MEF2C in the DDD cohort, but these would all be missed in standard clinical genetics approaches. Nonetheless, these variants are readily detectable in exome sequence data, with 30.7% of 5′ UTR bases across all genes well covered in the DDD dataset. Our analyses show that non-coding variants upstream of genes within which coding variants are known to cause DD are an important cause of severe disease and demonstrate that analyzing 5′ UTRs can increase diagnostic yield. We also show how non-coding variants can help inform both the disease-causing mechanism underlying protein-coding variants and dosage tolerance of the gene.
Keywords: developmental disorders, clinical genetic testing, non-coding region variants, 5' UTR variants
Introduction
The importance of non-coding regulatory variation in common diseases and traits has long been appreciated, but the contribution of non-coding variation to rare disease remains poorly understood.1, 2, 3, 4 Consequently, current clinical testing approaches for rare disease focus almost exclusively on regions of the genome that code directly for protein, within which we are able to relatively accurately estimate the effect of any individual variant. Using this approach, however, disease-causing variants are identified in only around 36% of individuals with developmental disorders (DD)5 using exome sequencing, with a further 15%–20% diagnosed through chromosomal microarrays.6 In previous work, we assessed the role of de novo mutations (DNMs) in distal regulatory elements and estimated that 1%–3% of undiagnosed individuals with DD carry pathogenic DNMs in these regions.1
Untranslated regions (UTRs) at the 5′ and 3′ end of genes present a unique opportunity to expand genetic testing outside of protein-coding regions given they have important regulatory roles in controlling both the amount and location of mRNA in the cell and the rate at which it is translated into protein.7,8 Crucially, we also know the genes/proteins that these regions regulate. Given that UTRs account for around the same genomic footprint as protein-coding exons, they have substantial potential to harbor novel Mendelian diagnoses.9,10 UTRs are, however, not regularly included in exome sequence capture regions and are excluded in most analysis pipelines. This is primarily due to a lack of guidance on how to determine when UTR variants are likely to be pathogenic.
Recently, we demonstrated that variants creating upstream start codons (uAUGs) in 5′ UTRs are under strong negative selection and are an important cause of Mendelian diseases, including neurofibromatosis and Van der Woude syndrome.11,12 Initiation of translation at a newly created uAUG can decrease translation of the downstream coding sequence (CDS). The strength of negative selection acting on uAUG-creating variants varies depending on both the match of the sequence surrounding the uAUG to the Kozak consensus, which is known to regulate the likelihood that translation is initiated,13,14 and the nature of the upstream open reading frame (uORF) that is created. Variants that result in ORFs which overlap the CDS have a larger impact on CDS translation and hence are more deleterious.11,15
Here, we screened 9,858 probands from the Deciphering Developmental Disorders (DDD)5 study for DNMs in the 5′ UTRs of genes within which variants have previously been shown to cause DD through a dominant haploinsufficient mechanism (defined using the clinically curated Developmental Disorders Genotype to Phenotype [DDG2P] database and henceforth referred to as “DDG2P haploinsufficient genes”). We uncover likely disease-causing variants that are entirely within non-coding regions and show how these variants cause disease through three distinct loss-of-function mechanisms. We further show how disease-causing missense variants in MEF2C (MIM: 600662) are clustered at the N terminus and likely also cause loss of function by disrupting binding of MEF2C protein to DNA. Finally, we analyze the coverage across all UTRs in the DDD exome sequencing dataset to demonstrate how these regions can be readily screened in existing datasets to increase diagnostic yield and glean insight into disease-causing mechanisms.
Material and methods
Recruitment, sample collection, and clinical data
The DDD Study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). Individuals with severe, undiagnosed developmental disorders and their parents were recruited and systematically phenotyped by the 24 Regional Genetics Services within the United Kingdom (UK) National Health Service and the Republic of Ireland. Saliva samples were collected from probands and parents, and DNA extracted as previously described;16 blood-extracted DNA was also collected for probands where available. Clinical data (growth measurements, family history, developmental milestones, etc.) were collected using a standard restricted-term questionnaire within DECIPHER.17 Informed consent was obtained for all participants.
Genetic data
Array-CGH analysis was performed using 2 × 1M probe custom designed microarrays (Agilent; Amadid No.s 031220/031221) as described previously.16 Exome sequencing was performed using Illumina HiSeq (75-base paired-end sequencing) with SureSelect baits (Agilent Human All-Exon V3 Plus and V5 Plus with custom ELID C0338371) and variants were called and annotated as described previously.16 We used DeNovoGear18 (v.0.54) to detect likely DNMs from trio exome BAM files and Ensembl Variant Effect Predictor19 was used to annotate predicted consequences. The data are available under managed access from the European Genome-phenome Archive (EGA: EGAS00001000775), and likely diagnostic variants are available open access in DECIPHER.
Defining a gene-set of interest
We limited our analysis to 359 DDG2P20 genes with a confirmed or probable role in developmental disorders and with a dominant (including X-linked dominant) loss-of-function disease mechanism (downloaded on 21st July 2020; see Web Resources section for link; Table S1). We refer to these genes as “DDG2P haploinsufficient genes.”
Identifying uAUG-creating variants in DDD
We defined high-confidence DNMs in DD as previously,21 using the following criteria: minor allele frequency < 0.01 in our cohort and reference databases, depth in the child > 7, depth in both parents > 5, Fisher strand bias p value > 10−3, and a posterior probability of being a DNM from DeNovoGear > 0.00781.18 Additionally, we filtered out DNMs with some evidence of an alternative allele in one of the parents and indels with a low variant allele fraction (<30% of the reads support the alternative) that had a minor allele frequency > 0. We cross-referenced this list of high-confidence DNMs with a list of all possible uAUG-creating SNVs from previous work.11 We also assessed any small insertions and deletions that could form uAUGs.
The strength of the Kozak consensus surrounding each uAUG was assessed as described previously.11 Specifically, we assessed the positions at −3 and +3 relative to the A of the AUG, requiring both the −3 base to be either A or G and the +3 to be G for an annotation of “strong.” If only one of these conditions was true, the strength was deemed to be “moderate” and if neither was the case “weak.”
Defining the 5′ UTR of MEF2C
We used the MANE Select transcript ENST00000504921.7 for which the 5′ UTR was defined using CAGE data from the FANTOM5 project,22 RNA-seq supported intron data from the Intropolis resource,23 and exon level expression from the GTEx project24. The Matched Annotation from the NCBI and EMBL-EBI (MANE) is a collaborative project that aims to define a representative transcript (MANE Select) for each protein-coding locus across the genome. The MANE set perfectly aligns to the GRCh38 reference assembly and includes pairs of 100% identical RefSeq and Ensembl/GENCODE transcripts.25 The 5′ UTR of MEF2C was therefore defined as two exons: chr5:88,178,772–88,179,001 and chr5:88,119,606–88,119,747 on GRCh37 or chr5:88,882,955–88,883,184 and chr5:88,823,789–88,823,930 on GRCh38.
Searching for MEF2C 5′ UTR variants in external datasets
We queried the regions corresponding to the MEF2C 5′ UTR for DNMs in (1) a set of 18,789 DD trios sequenced by the genetic testing company GeneDx,5 (2) 13,949 rare disease trios from the main program v9 release of the UK 100,000 Genomes Project from Genomics England,26 and (3) variants in the v3.0 dataset of the Genome Aggregation Database (gnomAD).27
Assessing 5′ UTR coverage
Regions corresponding to 5′ UTRs were extracted from the .gff file from the MANE project v.0.91 (see Web Resources; MANE Select transcripts). For each base, we calculated the mean coverage across 1,000 randomly selected samples from DDD. A mean coverage of >10× was used to call a base “covered.” Analysis was limited to genes with a defined MANE Select transcript. For our DDG2P haploinsufficient genes this was 345/359 genes (96.1%).
To identify all possible uAUG-creating variants in DDG2P haploinsufficient genes, we extracted the 5′ UTR sequence from the MANE rna.fna file and used the UTRannotator28 to find all possible uAUG-creating sites and annotate their consequence.
Functional validation of variants creating out-of-frame ORFs (oORFs): By MEF2C 5′ UTR-luciferase translation assay
Expression constructs
WT and variant MEF2C 5′ UTRs were cloned directly upstream of Gaussia luciferase (GLuc) in the pEZX-GA02 backbone (Labomics) and sequenced to confirm integrity. Secreted alkaline phosphatase (SEAP) was expressed on the same construct for normalization of transfection efficiency.
Cull culture, transfection, and analysis
HEK293T cells were purchased from ATCC and cultured in Dulbecco’s Modified Eagle Medium (glutamine+, pyruvate+) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were transfected with MEF2C 5′ UTR-luciferase constructs using Lipofectamine 3000, following manufacturer’s protocols. After 24 h, culture medium was sampled and GLuc and SEAP were simultaneously quantified using the Secrete-Pair Dual Luminescence assay (Genecopoeia). Fifteen technical replicates were performed across three independent experiments.
qPCR
RNA was purified from cells using phenol-chloroform extraction and the QIAGEN RNeasy Miniprep kit. RNA quantity was normalized and cDNA generated using IV VILO reverse transcriptase following manufacturer’s protocols. Quantitative PCR was performed using SYBR green master mix on a Quantstudio 7 Real-time PCR system and results normalized to co-amplified GAPDH. The following primers were used: GLUC F: 5′-CTGTCTGATCTGCCTGTCCC-3′, GLUC R: 5′-GGACTCTTTGTCGCCTTCGT-3′, SEAP F: 5′-ACCTTCATAGCGCACGTCAT-3′ and SEAP R: 5′-TCTAGAGTAACCCGGGTGCG-3′, GAPDH F: 5′-GGAGTCAACGGATTTGGTCG-3′, GAPDH R: ATCGCCCCACTTGATTTTGG-3′.
Kozak mutagenesis
The Kozak context of the c.−103G>A MEF2C 5′ UTR-luciferase construct was modified using the Quikchange II mutagenesis kit, following manufacturers protocols. The following PAGE-purified mutagenesis primers were used: F: 5′-CTCCTTCTTCAGCATTTTCACAGCTCAGTTCCCAA-3′, R: 5′-TTGGGAACTGAGCTGTGAAAATGCTGAAGAAGGAG-3′. Constructs were fully sequenced to verify mutation and construct integrity in each case
Functional validation of CDS-elongating variants: By MEF2 binding site-luciferase transactivation assay
Expression and reporter constructs
WT and variant MEF2C 5′ UTR+CDS oligos were cloned into the pReceiver-M02 expression construct (Labomics) and sequenced to confirm integrity. For normalization of transfection efficiency, cells were co-transfected with pRL-Renilla. A desMEF2-luciferase reporter construct was used to quantify the transactivational efficiency of each MEF2C expression construct, and consisted of three copies of a high-affinity MEF2 binding site,29 linked to an hsp68 minimal promoter in pGL3 (Promega).30
Cell culture and transfection
HL1 cardiomyocytes were cultured in Claycomb medium, supplemented with 2 mM L-glutamine, 10% FBS, and 100 g/mL penicillin/streptomycin. Culture surfaces were pre-treated with gelatin/fibronectin. Cells were co-transfected with (1) desMEF2-luciferase reporter construct, (2) pRL-Renilla transfection control, and (3) expression construct of either: (a) empty pcDNA3.1 (negative control), (b) WT MEF2C 5′ UTR+CDS, (c) MEF2C −26C>T, or (d) MEF2C −8C>T. Transfection was with Lipofectamine 2000, following manufacturers protocols. 48 h after transfection, firefly and Renilla Luciferases were quantified by the Promega Dual-Luciferase Reporter Assay System. Eighteen technical replicates were performed across three independent experiments.
Western blot
HL1 cells were lysed in RIPA buffer in the presence of protease and phosphatase inhibitors (04693159001 and 04906845001, Roche Diagnostics). Lysates were separated on SDS-PAGE gels and transferred to PVDF membranes, which were blocked with 3% skimmed milk in TBS. The primary antibody was anti-MEF2C (ab211493, Abcam), and the secondary antibody was anti-mouse P0447 from Dako. The membrane was developed using ECL reagent (AC2204, Azure Biosystems) and intensity of the bands quantified using ImageJ software.
Statistical analysis for all assays
Data were analyzed for statistical significance using 1-way ANOVA followed by Tukey’s post-test, using GraphPad Prism 8.0.
CNV calling
Four CNV detection algorithms (XHMM,31 CONVEX,16 CLAMMS,32 and CANOES33) were used to ascertain CNVs from exome data, followed by a random forest machine learning approach to integrate and filter the results.
Layered H3K4me3 data (to visualize active promoter regions) was downloaded from the UCSC table browser for GN12878 as a representative cell line and plotted alongside the identified CNVs in Figure S1.
Modeling missense disruption to DNA-binding
We collated a set of missense variants identified in MEF2C in DD-affected individuals comprising all de novo variants from trios in DDD and GeneDx published previously,5 and variants from ClinVar either flagged as being identified as de novo, or with functional evidence (Table S3).
As a comparator, we used missense variants from gnomAD v.2.1.1.27 Given that there are only three variants in the N-terminal region of MEF2C in gnomAD but the sequence of the N-terminal region is near identical across the four MEF2 proteins (Figure S4), we used missense variants from all four genes (MEF2A-D; Table S4).
Based on structures of the N-terminal MADS-box of MEF2A homodimer (PDB: 1EGW, 3KOV, and 6BYY, residues 1–92) bound to its DNA consensus sequence,34 we categorized residues into one of four categories: (1) in N-terminal random coil and in contact with the DNA; (2) in N-terminal alpha-helix pointing toward the DNA; (3) in N-terminal alpha-helix pointing away from the DNA; or (4) distal to the DNA contact surface (Table S5). We used a two-sided Fisher’s exact test to assess for an enrichment of variants in contact or pointing toward the DNA helix in DD-affected individuals (Table S6).
The Swissmodel threaded model of MEF2C based upon PDB: 6BYY (89% identity)35,36 was energy minimized using Pyrosetta37 with 15 FastRelax cycles38 against the electron density of PDB: 6BYY and 5 unconstrained. The DNA was extended on both ends due to the proximity of R15. Mutations were introduced and the 10 Å neighborhood was energy minimized. Gibbs free energy was calculated using the Rosetta ref2015 scorefunction.39 Gibbs free energy of binding was calculated by pulling away the DNA and repacking sidechains and, in the case of residues in the N-terminal loop, thoroughly energy minimizing the backbone of the loop as this is highly flexible when unbound. N-terminal extensions were made using the RemodelMover40 with residues 2–5 also remodelled as determined by preliminary test. Closest distance of each residue to the DNA was calculated with the Python PyMOL module. Code used for this analysis can be found at the link in Web Resources. This interactive page was made in Michelaɴɢʟo.41
All missense variants are annotated with respect to the Ensembl canonical transcript ENST00000340208.5.
Calculating regional missense constraint and de novo enrichment
We determined regional missense constraint by (1) extracting observed variant counts from the 125,748 samples in gnomAD v.2.1.1, (2) calculating the expected variant count per transcript, and (3) applying a likelihood ratio test to search for significant breaks that split a transcript into two or more sections of variable missense constraint.
Observed missense variants were extracted from the gnomAD exomes Hail Table (v.2.1.1) as described previously,27 using the following criteria:
-
•
Annotated as a missense change in a canonical transcript of a protein-coding gene in Gencode v.19 by Variant Effect Predictor (VEP, v.85)
-
•
Median coverage greater than zero in the gnomAD exomes data
-
•
Passed variant filters
-
•
Adjusted allele count of at least one and an allele frequency less than 0.1% in the gnomAD exomes
To calculate the expected variant count, we extended methods described previously27 to compute the proportion of expected missense variation per base. Briefly, we annotated each possible substitution with local sequence context, methylation level (for CpGs), and associated mutation rate from the table computed in Karczewski et al.27 We aggregated these mutation rates across the transcript and calibrated models based on CpG status and median coverage. To determine the expected variants for a given section of the transcript, we calculated the fraction of the overall the mutation rate represented by the section and multiplied it by the aggregated expected variant count for the full transcript.
We defined missense constraint by extending the methods from Samocha et al.42 We employed a likelihood ratio test to compare the null model (transcript has no regional variability in missense constraint) with the alternative model (transcript has evidence of regional variability in missense constraint). We required a χ2 value above a threshold of 10.8 to determine significance for each breakpoint, and in the case of multiple breakpoints, retained the breakpoint with the maximum χ2. This approach defined a single breakpoint in the MEF2C canonical transcript at chr5:88,057,138 (GRCh37).
To evaluate the enrichment of DNMs in the transcript when removing the N-terminal section, we determined the probability of a missense mutation in that region and then compared the observed number of DNMs (n = 3) with the expected count in 28,641 individuals using a Poisson test. Specifically, we took the probability of a missense mutation (mu_mis) as provided in the gnomAD v.2.1 constraint files for MEF2C and adjusted it for the fraction of mutability represented in the latter section of the gene (∼79.5%).
Results
Identifying de novo 5′ UTR variants in individuals with DD
To investigate the contribution of uAUG-creating variants to severe DD, we analyzed 29,523 high-confidence DNMs identified in exome sequencing data from 9,858 parent-offspring trios in the DDD study.5 Although the majority of DNMs identified are coding, as expected with exome sequencing data, many non-coding variants are also detectable, particularly near exon boundaries. Given that uAUG-creating variants that decrease CDS translation would only be expected to be deleterious in genes that are dosage sensitive, we restricted our analysis to the 5′ UTRs of 359 haploinsufficient genes from the curated DDG2P database20 (Table S1).
We identified five unique uAUG-creating de novo single-nucleotide variants (SNVs) in five unrelated probands upstream of two different genes. All of these variants are absent from the Genome Aggregation Database (gnomAD) population reference dataset (both v.2.1.1 and v.3.0).27 Notably, four of the five variants were found in the 5′ UTR of MEF2C in probands with phenotypes consistent with MEF2C haploinsufficiency (Table 1; [MIM: 613443]).43 Two of these DNMs create uAUGs out-of-frame with the MEF2C CDS, which are expected to reduce downstream protein translation, while the other two create uAUGs in-frame with the CDS, which are expected to elongate the protein (Figure 1). The fifth variant was located in a strong Kozak consensus upstream of STXBP1 (ENST00000373302.8:c.−26C>G), creating an uAUG out-of-frame with the STXBP1 CDS; the phenotype of the proband with this variant is consistent with STXBP1 haploinsufficiency,44 including global developmental delay, microcephaly, and delayed speech and language development.
Table 1.
Details of MEF2C uAUG-creating and upstream deletion variants discussed in this work
| Variant (GRCh37) | cDNA description (ENST00000504921.7) | Variant effect | Deletion size | Kozak strength | Proband ID(s) | Proband count | gnomAD v3 AC |
|---|---|---|---|---|---|---|---|
| uUAG-creating de novo variants discovered in probands with DD | |||||||
| chr5:88,119,671 T>A | c.−66A>T | out-of-frame oORF created | – | moderate | 1 | 1 | – |
| chr5:88,119,708 C>T | c.−103G>A | out-of-frame oORF created | – | weak | 2 | 1 | – |
| chr5:88,119,613 G>A | c.−8C>T | CDS-elongating | – | strong | 3,4,5 | 3 | – |
| chr5:88,119,631 G>A | c.−26C>T | CDS-elongating | – | moderate | 6,7,8 | 3 | – |
| uAUG-creating variant present in gnomAD: | |||||||
| chr5:88,883,052 G>A | c.−240C>T | uORF created | – | weak | – | 0 | 1 |
| chr5:88,883,059 G>A | c.−247C>T | uORF created | – | weak | – | 0 | 6 |
| Non-coding CNVs | |||||||
| chr5:88,133,089–88,427,361del | – | promoter and partial 5′ UTR deletion | 294 kb | – | 9 | 1 | – |
| chr5:88,123,099–88,220,350del | – | promoter and partial 5′ UTR deletion | 97 kb | – | 10 | 1 | – |
Shown are the four uAUG SNVs identified in DDD, uAUG SNVs observed in gnomAD v3.0, and non-coding CNVs found upstream of MEF2C in DDD. oORF = overlapping ORF; uORF = upstream ORF; AC = allele count. Proband IDs refer to those used in Table S2.
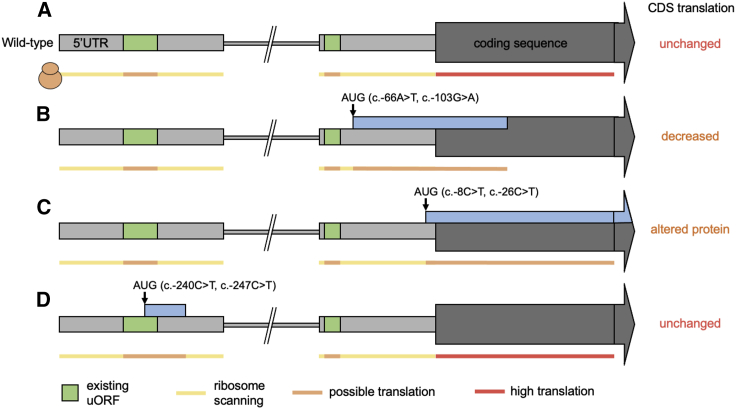
Figure 1.
Schematic of the N terminus of MEF2C
Shown are the wild-type structure (A) and the position and effect of uAUG-creating variants identified as de novo in developmental disorder cases (B and C) and in gnomAD population control subjects (D).
(A) The two 5′ UTR exons are shown as light gray boxes, separated by an intron shown as a thinner broken gray line. Upstream open reading frames (uORFs) already present in the sequence are shown in green. Variant positions are represented by arrows. New ORFs created by the variants are shown as blue boxes.
(B) Two case variants create ORFs that overlap the coding sequence (CDS) out-of-frame (oORF-creating). If translation initiates at the uAUG, the ribosome will not translate the CDS.
(C) Two recurrent case variants create uAUGs in-frame with the CDS. If translation initiates at this uAUG, an elongated protein will be translated.
(D) Two variants identified in gnomAD create uORFs far upstream of the CDS which would not be predicted to disrupt translation of the normal protein.
Given the identification of multiple uAUG-creating de novo SNVs in MEF2C in the DDD study, we subsequently queried high-confidence DNMs identified in 18,789 trios with DD that were exome sequenced by GeneDx5 for additional MEF2C DNMs. We uncovered three additional de novo occurrences of two of the uAUG-creating variants observed in the DDD study. In addition, we identified a further de novo occurrence of one of these variants in a DD proband in the UK 100,000 Genomes Project26 (Table 1).
In a separate analysis, we analyzed copy-number variants (CNVs) identified in the DDD study using exome sequencing data and identified five de novo CNVs overlapping MEF2C (Figure S1). Two of these CNVs (each found in a single additional proband) overlap the 5′ UTR of MEF2C without impacting any of the coding exons (Table 1). These two non-coding CNVs delete the first exon of the MEF2C 5′ UTR and >40 kb of immediately upstream sequence (294 kb and 97 kb, respectively), removing the entire promoter (as defined by the Ensembl regulatory build45 and H3K4me3 peaks from ENCODE46) and likely abolishing transcription of this allele (Figure S1). There are no large deletions (>600 bps) in this upstream region in the gnomAD structural variant dataset (v.2.1).47 Both coding MEF2C disruptions and non-coding deletions further upstream of MEF2C that are predicted to disrupt enhancer function have been identified in DD probands previously.48,49
De novo 5′ UTR variants cause phenotypes consistent with MEF2C haploinsufficiency
We collated all available clinical data for the ten probands with MEF2C 5′ UTR de novo variants and in each case the observed phenotype is consistent with previously reported MEF2C haploinsufficiency50,51 (Table S2). Specifically, of the nine individuals for which detailed phenotypic information was available, the following features were noted: global developmental delay (9/9) with delayed or absent speech (9/9), seizures (8/9), hypotonia (5/9), and stereotypies (2/9). These probands had no other likely disease-causing variants in the coding sequence of MEF2C, or in any other DDG2P genes following exome sequencing.
uAUG-creating SNVs cause loss of function by reducing translation or disrupting protein function
The four uAUG-creating SNVs identified in MEF2C result in two different downstream effects. We used two distinct experimental approaches to evaluate the impact of (1) out-of-frame uAUG-creating variants on downstream translation and (2) CDS-elongating variants on MEF2C-dependent transactivation.
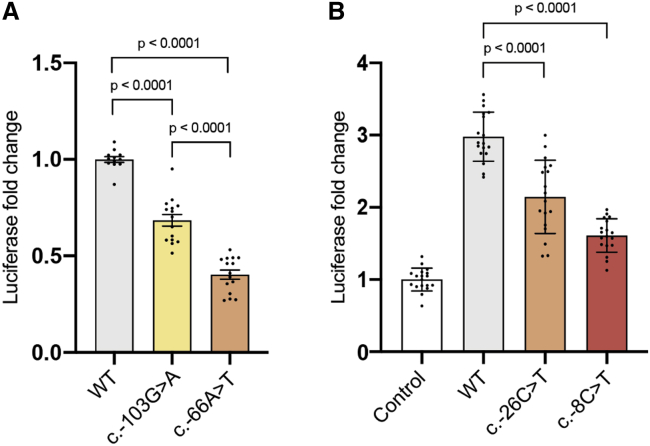
Two of the variants (c.−66A>T and c.−103G>A), each found in a single proband, create uAUGs that are out-of-frame with the coding sequence (CDS), creating an overlapping ORF (oORF) that terminates 128 bases after the canonical start site (Figure 1B). Using a translation assay, with wild-type or mutant 5′ UTR sequence cloned upstream of a luciferase reporter gene, we show that both variants result in a significant decrease in translational efficiency (Figures 2A and S2A). The amount by which translation is reduced appears to be dependent on the uAUG match to the Kozak consensus sequence, consistent with previous observations.11 The c.−103G>A variant, which creates an uAUG with a weak Kozak consensus, results in only a moderate decrease in luciferase expression, and the proband with this variant displays a milder phenotype on clinical review. To validate that this difference in effect is indeed due to the differing Kozak strengths, in the c.−103G>A translation assay, we mutated a single base to alter the oORF start context to a moderate Kozak consensus match (see material and methods). This modification resulted in significantly decreased translational efficiency compared to the unmodified c.−103G>A variant, to a level equivalent to the c.−66A>T variant (Figure S3). The individual carrying the c.−103G>A does not have any other 5′ UTR variants that could similarly modify the variant’s effect. These data suggest that MEF2C is sensitive to even partial loss of function.
Figure 2.
uAUG-creating variants decrease translation of MEF2C or transactivation of target genes
(A) MEF2C 5′ UTR out-of-frame overlapping ORF (oORF)-creating variants c.−103G>A and c.−66A>T (Figure 1B) reduce downstream luciferase expression relative to wild-type (WT) 5′ UTR in a translation reporter assay. Reduction is stronger for c.−66A>T (moderate uAUG Kozak context) than for c.−103G>A (weak Kozak context).
(B) Overexpression of MEF2C with the WT 5′ UTR/CDS induces expression of luciferase from a MEF2C-dependent enhancer-luciferase reporter construct, relative to an empty pcDNA3.1 construct negative control. The MEF2C N-terminus-extending variants c.−26C>T (9 amino acids) and c.−8C>T (3 amino acids; Figure 1C) both reduce transactivation.
Bars are colored by Kozak consensus: yellow = weak; orange = moderate; red = strong. Luciferase expression was normalized for transfection efficiency. Error bars represent the standard error of the mean. p values were calculated using 1-way ANOVA followed by Tukey’s post-test.
The other two variants (c.−8C>T and c.−26C>T) are both observed recurrently de novo, each in three unrelated probands (Table 1). Both variants create uAUGs that are in-frame with the CDS, resulting in N-terminal extensions of three and nine amino acids, respectively (Figure 1C). MEF2C is a transcription factor, and critical to its function is the DNA-binding domain located at the extreme N-terminal region.52 Although no structure is available for the MEF2C protein, numerous crystal and NMR structures of the N-terminal DNA-binding domain of human MEF2A are available, which is 96% identical in sequence to MEF2C. These structures show clearly that the extreme N terminus of the protein is in direct contact with DNA,34,53 and that the first few residues bind directly into the minor groove (Figure 3). We assayed MEF2C-dependent transactivation using MEF2C expression constructs with wild-type and mutant 5′ UTR sequences. These data demonstrate significantly reduced activation of target gene transcription from the variants (Figures 2B, S2B, and S2C) compared to wild-type MEF2C. Once again, the strength of the effect is dependent on the uAUG context, with the c.−8C>T variant that creates a strong Kozak consensus having a larger effect, almost abolishing transactivation activity.
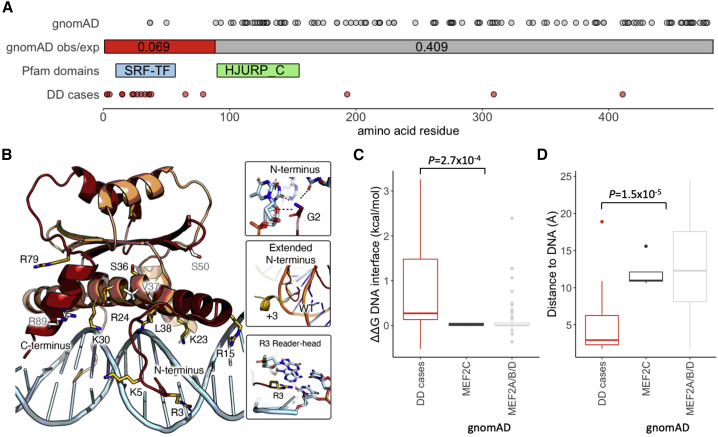
Figure 3.
MEF2C missense variants in individuals with DD are enriched in the N-terminal region and likely disrupt DNA binding
(A) The N-terminal region of MEF2C is highly constrained for missense variants in gnomAD (obs/exp = 0.069), with much lower constraint across the rest of the protein (obs/exp = 0.41). This region of high constraint correlates with the location of the majority of de novo missense variants identified in DD-affected individuals (red circles), while gnomAD variants are mostly outside of this N-terminal region (gray circles).
(B) The N-terminal portion of the MEF2C dimer (1–92), modeled using structures of the human MEF2A dimer which is 96% identical in sequence to MEF2C, bound directly to its consensus DNA sequence. Side chains of amino acids with pathogenic de novo missense variants from DDD, GeneDx, and ClinVar are shown in yellow, with gnomAD MEF2C missense variants in gray. Most pathogenic missense variants either protrude directly into the DNA or are located in the DNA-binding helix. In particular, the terminal amine (Gly2, top inset) along with Arg3 (bottom inset) act as reader-heads for nucleobase specificity, which is likely disrupted in the N-terminal extension variants (middle inset). All pathogenic and gnomAD variants can be viewed in our interactive protein structure browser (see link in Web Resources).
(C and D) Missense variants from DD-affected individuals (DDD, GeneDx, and ClinVar) are significantly more disruptive to the interaction with DNA as measured by ΔΔG values (C) and closer to the bound DNA molecule (D) than MEF2A-D variants in gnomAD (see STAR Methods). p values were calculated using a Wilcoxon rank sum test.
We looked in the gnomAD dataset27 for uAUG-creating variants that might have similar impacts. Across the exome (v.2.1.1) and genome (v.3.0) sequencing datasets, there are only two uAUG-creating variants in the MEF2C 5′ UTR. Crucially, neither of these fall into the proximal 5′ UTR exon and neither create ORFs overlapping the CDS. In both instances, the uAUGs are created into weak Kozak-consensus contexts, and they have in-frame stop codons after 6 bps (allele count = 6) and 57 bps (allele count = 1), respectively (Table 1; Figure 1D). These variants would therefore not be expected to have substantial, if any, effect on MEF2C translation.
Pathogenic de novo missense variants likely cause loss of function of MEF2C through disrupting DNA binding
While the major recognized mechanism through which pathogenic variants in MEF2C lead to severe developmental phenotypes is loss of function, de novo missense variants are also significantly enriched in DD trios (p = 1.3 × 10−14)5 and multiple pathogenic missense variants are reported in ClinVar.54 These variants are almost exclusively found at the extreme N terminus of the protein (Table S3), in the DNA-binding region, which is also highly constrained for missense variants in gnomAD (obs/exp = 0.069; calculated on 125,748 exome sequenced samples in v.2.1.1; Figure 3A). We hypothesized that these pathogenic missense variants are also causing loss of function by disrupting DNA binding of MEF2C as has been demonstrated for random disruptions to the N-terminal region52 and two proband variants49 previously. Using the structure of the N-terminal MEF2A homodimer bound to DNA, we modeled the location of pathogenic missense variants in MEF2C, as well as missense variants in gnomAD v.2.1.1 across all members of the myocyte enhancer factor 2 protein family (MEF2A-D; 84% N-terminal domain sequence identity; Table S4; Figure S4) and saw a significant enrichment of pathogenic variants interacting directly with DNA via both the N-terminal loop and DNA-binding helix (Fisher’s p = 2.6 × 10−5, Figure 3B; Tables S5 and S6). We further calculated the change in Gibbs free energy (ΔΔG) of both the protein-DNA interaction and the complex stability for each missense change. Variants found in individuals with DD have significantly increased ΔΔG scores compared to gnomAD variants (Wilcoxon p = 2.7 × 10−4; Figure 3C) and are significantly closer to the bound DNA (Wilcoxon p = 1.5 × 10−5; Figure 3D; Table S7). Together, these data suggest that disease-causing missense variants in MEF2C act through a loss-of-function mechanism, as has been experimentally demonstrated for two proband variants previously.49 Indeed, excluding the N-terminal DNA-binding domain, the remainder of MEF2C shows much weaker constraint against missense variants in gnomAD (obs/exp = 0.41) and only nominal enrichment for de novo missense variants in individuals with DD (p = 0.041).
Disease-causing 5′ UTR variants can be detected in exome-sequencing data
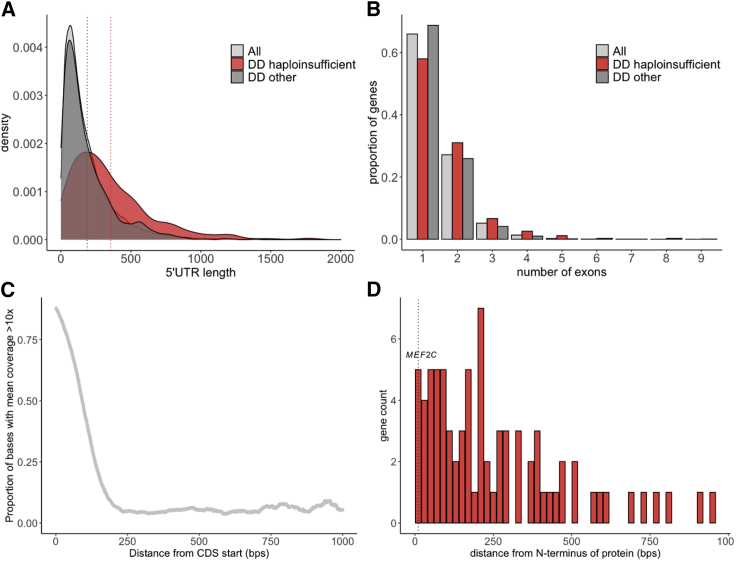
Given our ability to identify 5′ UTR variants in MEF2C, we investigated the extent to which these regions are captured across all genes in the exome sequencing dataset from the DDD study. We find that 30.7% of all gene 5′ UTR bases and 20.4% of 5′ UTR bases of our DDG2P haploinsufficient genes (average of 73 bps per gene; n = 345 with MANEv0.91 transcripts) are covered at a mean coverage threshold of >10×. The average length of 5′ UTRs in DDG2P haploinsufficient genes is 356 bps (Figure 4A), with 42.0% containing multiple exons (Figure 4B). As expected, 5′ UTR coverage decays as distance from the CDS increases (Figure 4C), with distal exons very poorly covered (6.7% of bases >10×). In comparison, a much lower proportion of 3′ UTR bases (6.0%) are covered at >10×, which is unsurprising given that 3′ UTRs are much longer than 5′ UTRs, at an average of 2,652 bps for our DDG2P haploinsufficient genes.
Figure 4.
Characteristics and sequencing coverage of the 5′ UTRs of DDG2P genes
(A and B) 5′ UTRs of DDG2P haploinsufficient genes (red) are longer (A), and a higher proportion have multiple exons (B) compared to 5′ UTRs of all genes (light gray), and other DDG2P genes (dark gray). Mean lengths for each gene set in (A) are shown as dotted lines.
(C) The coverage of 5′ UTRs decays rapidly with distance from the CDS (x axis truncated at 1,000 bps). Note that these figures were calculated using exome sequence data from the DDD study and may vary between different exome capture designs.
(D) The position of DNA-binding domains (including homeodomains, zinc-fingers, and specific DNA-binding domains) in DDG2P haploinsufficient genes with respect to the N terminus of the protein; MEF2C is one of three proteins with a DNA-binding domain that starts within 10 bps of the N terminus.
To determine the proportion of all possible uAUG-creating variants that are sufficiently covered in the DDD exome-sequence data, we computationally identified 3,962 possible uAUG-creating variants in DDG2P haploinsufficient genes that would create out-of-frame overlapping ORFs (n = 2,782) or CDS elongations (n = 1,180). Of these, 42.4% are sequenced at >10× coverage across the DDD study dataset (40.2% of out-of-frame and 47.6% of CDS elongating). However, we would not expect CDS-elongating variants to cause a loss of function for the majority of genes. Rather, we expect this to be limited to genes with important functional domains at the extreme N terminus that would be adversely affected by the addition of extra N-terminal amino acids, either through disrupting binding or altering protein structure. Based on Pfam domain predictions, only three of the proteins encoded by our 359 DDG2P haploinsufficient genes, including MEF2C, have DNA-binding domains that start within 10 bps of the Nterminus (Figure 4D); the other two (ZNF750 and SIM1) encode an N-terminal zinc-finger and basic helix-loop-helix, respectively, and although no structures are available, these bind DNA via specific motifs that are unlikely to include the extreme N-terminal residues.
Discussion
Here, we have identified six unique non-coding, pathogenic DNMs in MEF2C in ten individuals with severe developmental disorders (six in the DDD study, three in a cohort from GeneDx, and one in the UK 100,000 Genomes Project). These variants act via three distinct loss-of-function mechanisms at different stages of expression regulation: (1) two large deletions remove the promoter and part of the 5′ UTR and are predicted to abolish normal transcription of MEF2C; (2) two SNVs create out-of-frame uAUGs and reduce normal translation of the MEF2C coding sequence; and (3) two SNVs create in-frame uAUGs that elongate the MEF2C coding sequence, disrupting binding of the MEF2C protein to DNA and reducing subsequent transactivation of gene expression. We also identified a single uAUG-creating variant in STXBP1 in a proband whose phenotype was consistent with STXBP1 haploinsufficiency. This variant is predicted to create an out-of-frame oORF into a strong Kozak consensus, thus decreasing normal STXBP1 translation (as ribosomes first encounter and begin to translate from this new uAUG), leading to reduced levels of STXBP1 protein.
These observations demonstrate the importance of screening 5′ UTRs of genes known to harbor disease-causing coding variants in individuals that remain genetically undiagnosed. We have previously identified 20 probands with diagnostic DNMs (15 SNVs and 5 CNVs) impacting MEF2C protein-coding regions in the 9,858 family trios analyzed in the DDD study. The six additional non-coding DNMs described here (four SNVs and two CNVs) therefore comprise 23% of diagnoses impacting MEF2C in this cohort.
Our data show that 5′ UTR variants can be identified in existing datasets that were primarily designed to capture coding sequences, with 30.7% of 5′ UTR bases having sufficient (>10×) coverage in exome-sequencing data from the DDD study. However, exome-sequencing data is likely to only identify UTR variants that are proximal to the first and last exons of genes, and whole-genome or expanded panel sequencing will be required to assay distal or poorly covered UTRs. Furthermore, given their large size, 3′ UTRs are particularly poorly covered in exome-sequencing datasets. There are examples of disease-causing variants within 3′ UTRs, including those impacting poly(A) signals and microRNA binding,9,55, 56, 57 which will not be detected using these methodologies but that could increase diagnostic yield.
Although we screened DNMs in the 5′ UTRs of a set of 359 haploinsufficient DDG2P genes, four of the five identified de novo uAUG-creating variants were found in MEF2C. This enrichment in a single gene is likely due to a combination of factors (Figure S5). First, MEF2C has a proximal 5′ UTR exon that is very well covered in the DDD exome sequencing data. Second, this 5′ UTR exon contains a large number of sites where a variant could create an uAUG, with only two DDG2P haploinsufficient genes having more well-covered possible uAUG-creating sites. Third, unlike the other genes with well-covered possible uAUG-creating sites, MEF2C haploinsufficiency is a recurrent cause of DD within the DDD study (Figure S5). Finally, due to the direct interaction of the extreme N terminus of MEF2C with DNA, CDS-elongating variants are also likely to be pathogenic, which is unlikely to be the case in the vast majority of other haploinsufficient genes. As a result, MEF2C may be unusual in its potential for pathogenic mutations in the 5′ UTR and similarly large increases in diagnostic yield are unlikely across most DDG2P haploinsufficient genes. Nevertheless, the enrichment of uAUG-creating variants in MEF2C is striking: only 14 of 426 possible variants create uAUGs (at 142 5′ UTR bases that are well covered in the DDD study exome-sequencing data), yet all four DNMs observed in the DDD study in the MEF2C 5′ UTR are uAUG creating (binomial p = 1.2 × 10−6).
In our functional data, we see a difference in the size of variant effects dependent on the strength of the Kozak consensus surrounding the newly created uAUG. The Kozak sequence is known to influence the likelihood of a ribosome initiating translation at any given AUG as it scans along the 5′ UTR from the 5′ cap.13 Our four uAUG-creating variants each generate a new uORF that overlaps the coding sequence. Ribosomes that initiate translation at these uAUGs will not be available to translate from the wild-type coding start site (which itself has a strong Kozak consensus), resulting in reduced translation of the CDS. The stronger the Kozak consensus around the uAUG, the greater this effect will be.
As we extend our analyses to detect non-coding variants, we caution that interpretation of UTR variants still remains a critical challenge. Every 5′ UTR has a unique combination of regulatory elements tightly regulating RNA stability and protein expression,58,59 and the impact of any variant will vary with the gene-specific context. Functional validation of identified variants will therefore be crucial to prove (or reject) causality. Some variants may have only a partial regulatory effect, but these variants can nonetheless be harnessed to assess the extent to which perturbation of protein levels or function is tolerated, potentially leading to reduced expressivity and/or lower penetrance. In the case of MEF2C, our results suggest that even partial reductions in protein expression lead to severe disease.
Finally, we note how the mechanism of action of non-coding variants can inform the mechanisms underlying protein-coding variants. Identification and characterization of the effect of the CDS-elongating MEF2C variants led us to analyze the domain structure of MEF2C protein and confirm that all the currently identified missense variants likely also act via disrupting DNA binding, leading to a loss of function.
In conclusion, our results further highlight the important contribution of non-coding regulatory variants to rare disease and underscore the huge promise of large whole-genome-sequencing datasets to both find new diagnoses and further our understanding of regulatory disease mechanisms.
Declaration of interests
K.J.K. is a consultant for Vor Biopharma. J.J. and K.R. are employees of GeneDx, Inc. K.R. holds shares in Opko Health, Inc. B.D.Z. is a member of the speakers bureau for Biogen, Neurelis, and Supernus. S.A.C. is co-founder and shareholder of Enleofen Bio Pte Ltd. M.E.H. is co-founder, shareholder, consultant, and non-executive director of Congenica Ltd. All other authors declare no competing interests.
Acknowledgments
N.W. is currently supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (grant number 220134/Z/20/Z). Initial work was completed while N.W. was supported by a Rosetrees and Stoneygate Imperial College Research Fellowship. N.M.Q. is supported by the Imperial College Academic Health Science Centre. This work is additionally supported by The Rosetrees Trust (grant number H5R01320), the Wellcome Trust (WT200990/Z/16/Z, WT200990/A/16/Z), Fondation Leducq (16 CVD 03), the National Institute for Health Research (NIHR) Imperial College Biomedical Research Centre, the Cardiovascular Research Centre, Royal Brompton & Harefield NHS Trust, and the NIHR Oxford Biomedical Research Centre Programme. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003) a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute (grant number WT098051). See https://www.ddduk.org/access.html for full acknowledgment. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support. This research was funded, in whole or in part, by Wellcome. A CC BY or equivalent license is applied to the Author Accepted Manuscript, in accordance with Wellcome open access conditions.
Published: May 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.04.025.
Data and code availability
The DDD study data are available under managed access from the European Genome-phenome Archive (EGA: EGAS00001000775) and likely diagnostic variants are available open access in DECIPHER. Code used for modeling case and population variants on the MEF2C protein structure can be found here: https://github.com/matteoferla/MEF2C_analysis.
Web resources
Code for MEF2C protein modeling, https://github.com/matteoferla/MEF2C_analysis
European Genome-phenome Archive (EGA), https://www.ebi.ac.uk/ega
Gene-2-phenotype, https://www.ebi.ac.uk/gene2phenotype/downloads
Genomics England 100,000 Genomes Project de novo call set, https://cnfl.extge.co.uk/display/GERE/De+novo+variant+research+dataset
Interactive protein structure browser, https://michelanglo.sgc.ox.ac.uk/r/mef2c
Matched Annotation between NCBI and EBI project information (MANE), https://www.ncbi.nlm.nih.gov/refseq/MANE/
MANE data download, ftp://ftp.ncbi.nlm.nih.gov/refseq/MANE/MANE_human/release_0.91/
OMIM, https://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
UCSC table browser, https://genome.ucsc.edu/cgi-bin/hgTables
Supplemental information
References
- 1.Short P.J., McRae J.F., Gallone G., Sifrim A., Won H., Geschwind D.H., Wright C.F., Firth H.V., FitzPatrick D.R., Barrett J.C., Hurles M.E. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature. 2018;555:611–616. doi: 10.1038/nature25983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spielmann M., Mundlos S. Looking beyond the genes: the role of non-coding variants in human disease. Hum. Mol. Genet. 2016;25(R2):R157–R165. doi: 10.1093/hmg/ddw205. [DOI] [PubMed] [Google Scholar]
- 3.An J.-Y., Lin K., Zhu L., Werling D.M., Dong S., Brand H., Wang H.Z., Zhao X., Schwartz G.B., Collins R.L. Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science. 2018;362:362. doi: 10.1126/science.aat6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., Centers for Mendelian Genomics The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplanis J., Samocha K.E., Wiel L., Zhang Z., Arvai K.J., Eberhardt R.Y., Gallone G., Lelieveld S.H., Martin H.C., McRae J.F., Deciphering Developmental Disorders Study Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature. 2020;586:757–762. doi: 10.1038/s41586-020-2832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srivastava S., Love-Nichols J.A., Dies K.A., Ledbetter D.H., Martin C.L., Chung W.K., Firth H.V., Frazier T., Hansen R.L., Prock L., NDD Exome Scoping Review Work Group Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet. Med. 2019;21:2413–2421. doi: 10.1038/s41436-019-0554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mignone F., Gissi C., Liuni S., Pesole G. 2002. Untranslated regions of mRNAs. Genome Biol 3, reviews0004.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mortimer S.A., Kidwell M.A., Doudna J.A. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- 9.Wanke K.A., Devanna P., Vernes S.C. Understanding Neurodevelopmental Disorders: The Promise of Regulatory Variation in the 3'UTRome. Biol. Psychiatry. 2018;83:548–557. doi: 10.1016/j.biopsych.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee S., Pal J.K. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol. Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- 11.Whiffin N., Karczewski K.J., Zhang X., Chothani S., Smith M.J., Evans D.G., Roberts A.M., Quaife N.M., Schafer S., Rackham O., Genome Aggregation Database Production Team. Genome Aggregation Database Consortium Characterising the loss-of-function impact of 5′ untranslated region variants in 15,708 individuals. Nat. Commun. 2020;11:2523. doi: 10.1038/s41467-019-10717-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lima R.L.L.F., Hoper S.A., Ghassibe M., Cooper M.E., Rorick N.K., Kondo S., Katz L., Marazita M.L., Compton J., Bale S. Prevalence and nonrandom distribution of exonic mutations in interferon regulatory factor 6 in 307 families with Van der Woude syndrome and 37 families with popliteal pterygium syndrome. Genet. Med. 2009;11:241–247. doi: 10.1097/GIM.0b013e318197a49a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noderer W.L., Flockhart R.J., Bhaduri A., Diaz de Arce A.J., Zhang J., Khavari P.A., Wang C.L. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol. Syst. Biol. 2014;10:748. doi: 10.15252/msb.20145136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sample P.J., Wang B., Reid D.W., Presnyak V., McFadyen I.J., Morris D.R., Seelig G. Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nat. Biotechnol. 2019;37:803–809. doi: 10.1038/s41587-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramu A., Noordam M.J., Schwartz R.S., Wuster A., Hurles M.E., Cartwright R.A., Conrad D.F. DeNovoGear: de novo indel and point mutation discovery and phasing. Nat. Methods. 2013;10:985–987. doi: 10.1038/nmeth.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thormann A., Halachev M., McLaren W., Moore D.J., Svinti V., Campbell A., Kerr S.M., Tischkowitz M., Hunt S.E., Dunlop M.G. Flexible and scalable diagnostic filtering of genomic variants using G2P with Ensembl VEP. Nat. Commun. 2019;10:2373. doi: 10.1038/s41467-019-10016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M., Itoh M., FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nellore A., Jaffe A.E., Fortin J.-P., Alquicira-Hernández J., Collado-Torres L., Wang S., Phillips R.A., III, Karbhari N., Hansen K.D., Langmead B., Leek J.T. Human splicing diversity and the extent of unannotated splice junctions across human RNA-seq samples on the Sequence Read Archive. Genome Biol. 2016;17:266. doi: 10.1186/s13059-016-1118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium T.G., GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yates A.D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R. Ensembl 2020. Nucleic Acids Res. 2020;48(D1):D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caulfield M., Davies J., Dennys M., Elbahy L., Fowler T., Hill S., Hubbard T., Jostins L., Maltby N., Mahon-Pearson J. 2019. The National Genomics Research and Healthcare Knowledgebase.https://www.genomicsengland.co.uk/wp-content/uploads/2019/08/The-National-Genomics-Research-and-Healthcare-Knowledgebase-v5-1.pdf [Google Scholar]
- 27.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., Wakeling M., Ware J., Whiffin N. Annotating high-impact 5′untranslated region variants with the UTRannotator. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa783. Published online September 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naya F.J., Wu C., Richardson J.A., Overbeek P., Olson E.N. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development. 1999;126:2045–2052. doi: 10.1242/dev.126.10.2045. [DOI] [PubMed] [Google Scholar]
- 30.Wu H., Rothermel B., Kanatous S., Rosenberg P., Naya F.J., Shelton J.M., Hutcheson K.A., DiMaio J.M., Olson E.N., Bassel-Duby R., Williams R.S. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO J. 2001;20:6414–6423. doi: 10.1093/emboj/20.22.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fromer M., Moran J.L., Chambert K., Banks E., Bergen S.E., Ruderfer D.M., Handsaker R.E., McCarroll S.A., O’Donovan M.C., Owen M.J. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am. J. Hum. Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Packer J.S., Maxwell E.K., O’Dushlaine C., Lopez A.E., Dewey F.E., Chernomorsky R., Baras A., Overton J.D., Habegger L., Reid J.G. CLAMMS: a scalable algorithm for calling common and rare copy number variants from exome sequencing data. Bioinformatics. 2016;32:133–135. doi: 10.1093/bioinformatics/btv547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Backenroth D., Homsy J., Murillo L.R., Glessner J., Lin E., Brueckner M., Lifton R., Goldmuntz E., Chung W.K., Shen Y. CANOES: detecting rare copy number variants from whole exome sequencing data. Nucleic Acids Res. 2014;42:e97. doi: 10.1093/nar/gku345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santelli E., Richmond T.J. Crystal structure of MEF2A core bound to DNA at 1.5 A resolution. J. Mol. Biol. 2000;297:437–449. doi: 10.1006/jmbi.2000.3568. [DOI] [PubMed] [Google Scholar]
- 35.Bienert S., Waterhouse A., de Beer T.A.P., Tauriello G., Studer G., Bordoli L., Schwede T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017;45(D1):D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei X., Kou Y., Fu Y., Rajashekar N., Shi H., Wu F., Xu J., Luo Y., Chen L. The Cancer Mutation D83V Induces an α-Helix to β-Strand Conformation Switch in MEF2B. J. Mol. Biol. 2018;430:1157–1172. doi: 10.1016/j.jmb.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Chaudhury S., Lyskov S., Gray J.J. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics. 2010;26:689–691. doi: 10.1093/bioinformatics/btq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conway P., Tyka M.D., DiMaio F., Konerding D.E., Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2014;23:47–55. doi: 10.1002/pro.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alford R.F., Leaver-Fay A., Jeliazkov J.R., O’Meara M.J., DiMaio F.P., Park H., Shapovalov M.V., Renfrew P.D., Mulligan V.K., Kappel K. The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design. J. Chem. Theory Comput. 2017;13:3031–3048. doi: 10.1021/acs.jctc.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang P.-S., Ban Y.-E.A., Richter F., Andre I., Vernon R., Schief W.R., Baker D. RosettaRemodel: a generalized framework for flexible backbone protein design. PLoS ONE. 2011;6:e24109. doi: 10.1371/journal.pone.0024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferla M.P., Pagnamenta A.T., Damerell D., Taylor J.C., Marsden B.D. MichelaNglo: sculpting protein views on web pages without coding. Bioinformatics. 2020;36:3268–3270. doi: 10.1093/bioinformatics/btaa104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samocha K.E., Kosmicki J.A., Karczewski K.J., O’Donnell-Luria A.H., Pierce-Hoffman E., MacArthur D.G., Neale B.M., Daly M.J. Regional missense constraint improves variant deleteriousness prediction. bioRxiv. 2017 doi: 10.1101/148353. [DOI] [Google Scholar]
- 43.Le Meur N., Holder-Espinasse M., Jaillard S., Goldenberg A., Joriot S., Amati-Bonneau P., Guichet A., Barth M., Charollais A., Journel H. MEF2C haploinsufficiency caused by either microdeletion of the 5q14.3 region or mutation is responsible for severe mental retardation with stereotypic movements, epilepsy and/or cerebral malformations. J. Med. Genet. 2010;47:22–29. doi: 10.1136/jmg.2009.069732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suri M., Evers J.M.G., Laskowski R.A., O’Brien S., Baker K., Clayton-Smith J., Dabir T., Josifova D., Joss S., Kerr B., DDD Study Protein structure and phenotypic analysis of pathogenic and population missense variants in STXBP1. Mol. Genet. Genomic Med. 2017;5:495–507. doi: 10.1002/mgg3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zerbino D.R., Wilder S.P., Johnson N., Juettemann T., Flicek P.R. The ensembl regulatory build. Genome Biol. 2015;16:56. doi: 10.1186/s13059-015-0621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A., Kaul R., ENCODE Project Consortium Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583:699–710. doi: 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collins R.L., Brand H., Karczewski K.J., Zhao X., Alföldi J., Francioli L.C., Khera A.V., Lowther C., Gauthier L.D., Wang H., Genome Aggregation Database Production Team. Genome Aggregation Database Consortium A structural variation reference for medical and population genetics. Nature. 2020;581:444–451. doi: 10.1038/s41586-020-2287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D’haene E., Bar-Yaacov R., Bariah I., Vantomme L., Van Loo S., Cobos F.A., Verboom K., Eshel R., Alatawna R., Menten B. A neuronal enhancer network upstream of MEF2C is compromised in patients with Rett-like characteristics. Hum. Mol. Genet. 2019;28:818–827. doi: 10.1093/hmg/ddy393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zweier M., Gregor A., Zweier C., Engels H., Sticht H., Wohlleber E., Bijlsma E.K., Holder S.E., Zenker M., Rossier E. Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Hum. Mutat. 2010;31:722–733. doi: 10.1002/humu.21253. [DOI] [PubMed] [Google Scholar]
- 50.Bienvenu T., Diebold B., Chelly J., Isidor B. Refining the phenotype associated with MEF2C point mutations. Neurogenetics. 2013;14:71–75. doi: 10.1007/s10048-012-0344-7. [DOI] [PubMed] [Google Scholar]
- 51.Vrečar I., Innes J., Jones E.A., Kingston H., Reardon W., Kerr B., Clayton-Smith J., Douzgou S. Further Clinical Delineation of the MEF2C Haploinsufficiency Syndrome: Report on New Cases and Literature Review of Severe Neurodevelopmental Disorders Presenting with Seizures, Absent Speech, and Involuntary Movements. J. Pediatr. Genet. 2017;6:129–141. doi: 10.1055/s-0037-1601335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molkentin J.D., Black B.L., Martin J.F., Olson E.N. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol. 1996;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Potthoff M.J., Olson E.N. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 54.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bennett C.L., Brunkow M.E., Ramsdell F., O’Briant K.C., Zhu Q., Fuleihan R.L., Shigeoka A.O., Ochs H.D., Chance P.F. A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA-->AAUGAA) leads to the IPEX syndrome. Immunogenetics. 2001;53:435–439. doi: 10.1007/s002510100358. [DOI] [PubMed] [Google Scholar]
- 56.Devanna P., Chen X.S., Ho J., Gajewski D., Smith S.D., Gialluisi A., Francks C., Fisher S.E., Newbury D.F., Vernes S.C. Next-gen sequencing identifies non-coding variation disrupting miRNA-binding sites in neurological disorders. Mol. Psychiatry. 2018;23:1375–1384. doi: 10.1038/mp.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reamon-Buettner S.M., Cho S.-H., Borlak J. Mutations in the 3′-untranslated region of GATA4 as molecular hotspots for congenital heart disease (CHD) BMC Med. Genet. 2007;8:38. doi: 10.1186/1471-2350-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Araujo P.R., Yoon K., Ko D., Smith A.D., Qiao M., Suresh U., Burns S.C., Penalva L.O.F. Before It Gets Started: Regulating Translation at the 5′ UTR. Comp. Funct. Genomics. 2012;2012:475731. doi: 10.1155/2012/475731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leppek K., Das R., Barna M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018;19:158–174. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DDD study data are available under managed access from the European Genome-phenome Archive (EGA: EGAS00001000775) and likely diagnostic variants are available open access in DECIPHER. Code used for modeling case and population variants on the MEF2C protein structure can be found here: https://github.com/matteoferla/MEF2C_analysis.