Figure 3.
Zebrafish ipo8 mutants show a range of dorsalization phenotypes
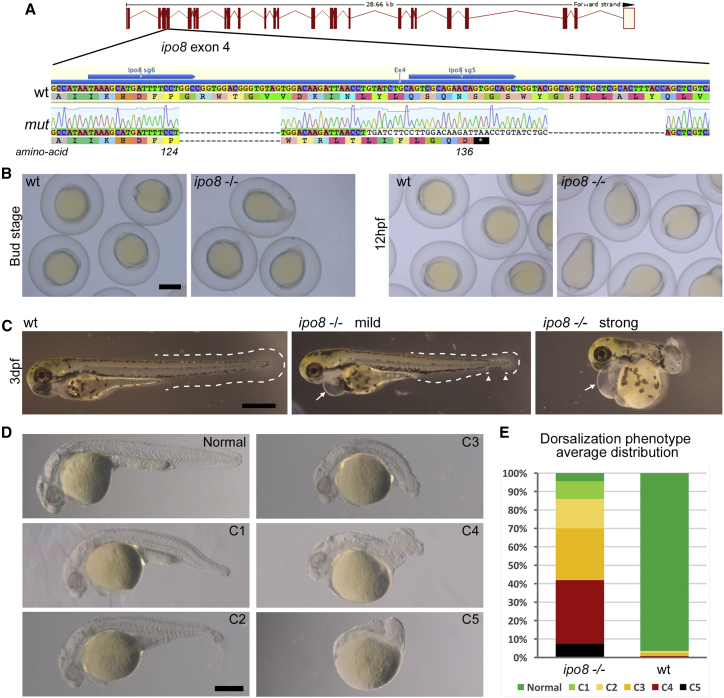
(A) Schematics of CRISPR/Cas9-mediated gene disruption at the ipo8 genomic locus. The sgRNAs (sg5 and sg6, blue arrows) targeted exon 4. Compared to the wild-type (WT) sequence, the mutated allele from the founder fish (mut) displayed two deletions of 18 and 22 bases and an insertion of 34 bp generating a frameshift from amino acid 124 and a premature STOP codon after 136 amino acids.
(B) Bright field pictures of bud (10 hpf) and early somite stages (12 hpf) embryos showing the elongating shape of the ipo8−/− embryos compared to WT controls. Scale bar represents 500 μm.
(C) Bright field pictures of 3 dpf larvae presenting the variable penetrance of the ipo8−/− phenotype (mild and strong) compared to WT controls. The tail fin is circled with dashed lines when visible. Gaps are highlighted by arrowheads. Arrows point at heart edema. Scale bar represents 200 μm.
(D) Nomarski pictures depicting the different classes of the dorsalization phenotype at 24 hpf as described in Mullins et al.26 (from severe C5 to mild C1) in the ipo8−/− mutants compared to the normal phenotype of a WT embryo. Scale bar represents 200 μm.
(E) Quantification of the distribution of the dorsalization classes in ipo8−/− mutants (5 clutches, n = 903) compared to WT clutches (4 WT clutches, n = 503). Average values in ipo8−/− mutants were 7.3% C5, 34.6% C4, 28.0% C3, 16.0% C2, 9.6% C1, and 4.5% normal, and average values in WT controls were 0% C5, 0.8% C4, 1.6% C3, 1.1% C2, 0% C1, and 96.5% normal.