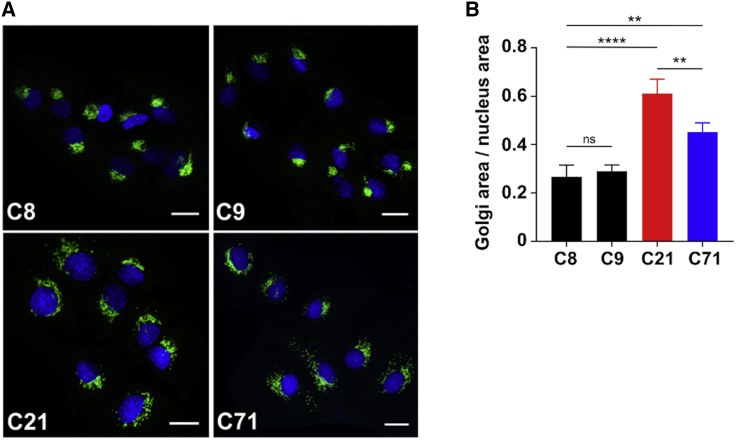
Figure 5.
Abnormal Golgi structure and function in p.Arg423∗ base-edited Huh7 cells
(A) Immunofluorescence staining of Huh7 control (C8 and C9) and edited cells (C21 and C71) with the Golgi marker, GM130, showing abnormalities Golgi morphology/area. Scale bar represents 20 µm.
(B) Golgi area was quantified and then normalized to the area of the nucleus to provide a ratio showing both C21 (homozygous clone) and C71 (heterozygous clone) had significantly increased Golgi area. The effect is significantly more pronounced when both alleles are mutated as they are in C21. In three separate biological replicates, conducted within monthly intervals, each time with freshly thawed cells, n = 8 cells were analyzed, and the mean was taken. The graph represents an average of the means acquired over three different biological measurements. Total cells measured N = 24 over three biological replicates. Statistical significance ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 was calculated via one-way ANOVA.