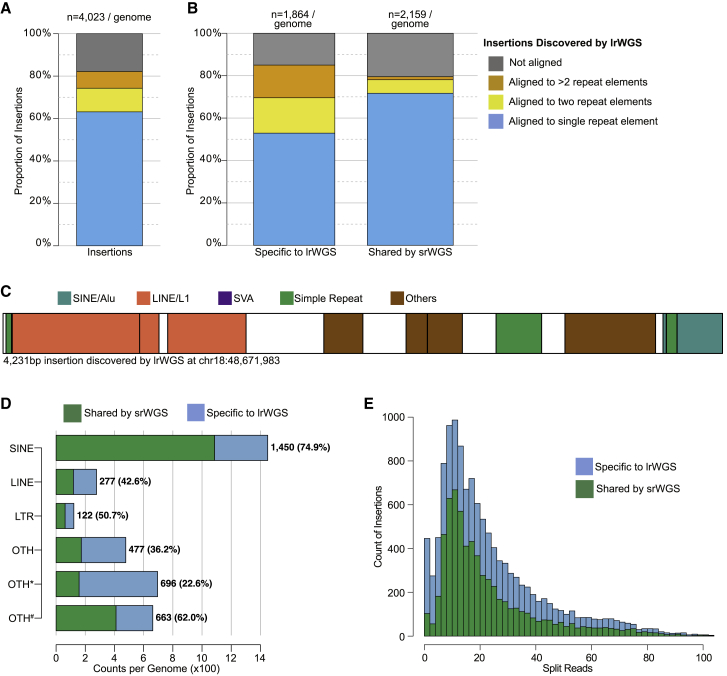
Figure 3.
Alignment of assembled lrWGS insertion sequences against known repeat elements
(A) Count of lrWGS insertions in "unique + RM" sequences per genome by alignment of inserted sequences to known repeat elements. The number on top of the bar represents the averaged count of high-confidence insertions in "unique + RM" sequences per genome.
(B) Count of lrWGS insertions that are specifically discovered by lrWGS and shared by srWGS, by alignment of inserted sequences to known repeat elements. Formatting conventions are the same as in (A).
(C) An example of an insertion SV assembled by lrWGS, annotated with sequences that align to known repeat element classes. White shading represents sequences not annotated as a known repeat element.
(D) Counts of lrWGS insertions in "unique + RM" sequences per genome by the class of inserted sequence and the proportion that was overlapped by srWGS. “OTH∗” represents insertions aligned to multiple known repeat elements, such as the example shown in (B). “OTH#” stands for insertions that were not aligned to any repeat elements. Numbers in parentheses represent the proportion of insertions that were overlapped by srWGS.
(E) Count of split reads around the lrWGS high-confidence insertions in histogram.