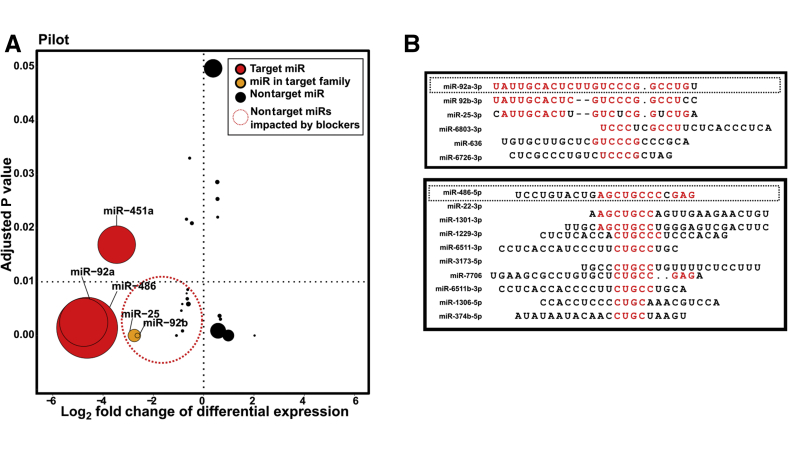
Figure 4.
Some nonheme microRNAs (miRNAs) are reduced by blocking oligonucleotides and share modest sequence similarity with target miRs A: Graph summarizing factors involved in determining true off-targets (see Table 1 for values). All miRs to the right of the vertical dotted line (positive fold change) were significantly increased in blocked libraries. All miRs below the horizontal dotted line were significantly (adjusted P < 0.01) differentially expressed. Red points are targets miRs, and orange points are miRs in the same family as a target miR. Size of point scales with the total number of mean counts per million that support the miRNA. The dotted circled portion denotes miRNA species that may be of concern as off-target effects. B: Sequence similarity of potential off-target effects with target sequences. 3p/5p designation added based on which version of the miR shared sequence similarity with a target miR. Red text denotes regions of alignment between target heme miR and off target miR. Dotted boxes denote sequences of miR92a and miR-486 used for blocking oligonucleotides.