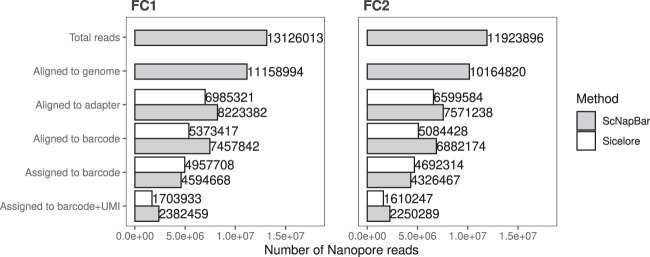
FIGURE 4.
Number of the Nanopore reads identified by ScNapBar and Sicelore at each processing step. We inspected each processing step on real data (low lllumina saturation of 11.3%). The first two steps are identical for both workflows. Total Reads: Number of input reads, aligned to genome: Number of reads aligned to genome. The next three steps are workflow-specific: Aligned to adapter: Number of reads with identified adapter sequence, aligned to barcode: Number of reads with aligned barcode sequence, Assigned to barcode: Number of predictions by each workflow. The last step is a validation of the previous assignment step after additional Illumina sequencing, which increases the Illumina saturation to 52%, and using UMI matches, see main text.