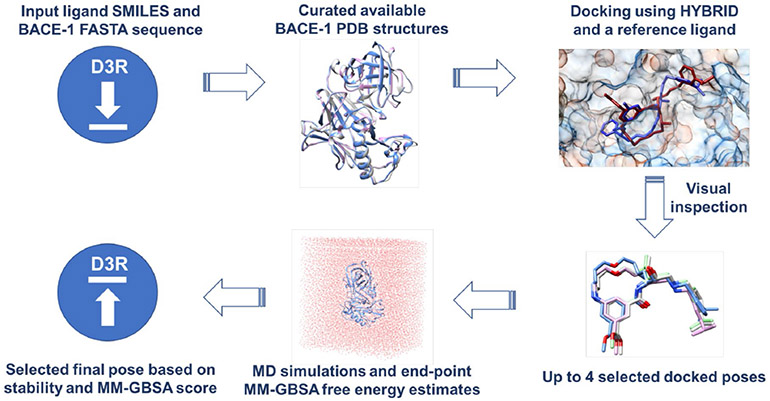
Fig. 2.
A schematic description of our ligand similarity-based protocol used for pose prediction in Stage 1a. For Stage 1b, we directly used the receptor structures provided by the organizers to dock the ligands using HYBRID. The down arrow represents the input files provided by the D3R GC4 organizers and the up arrow represents our pose prediction submissions.