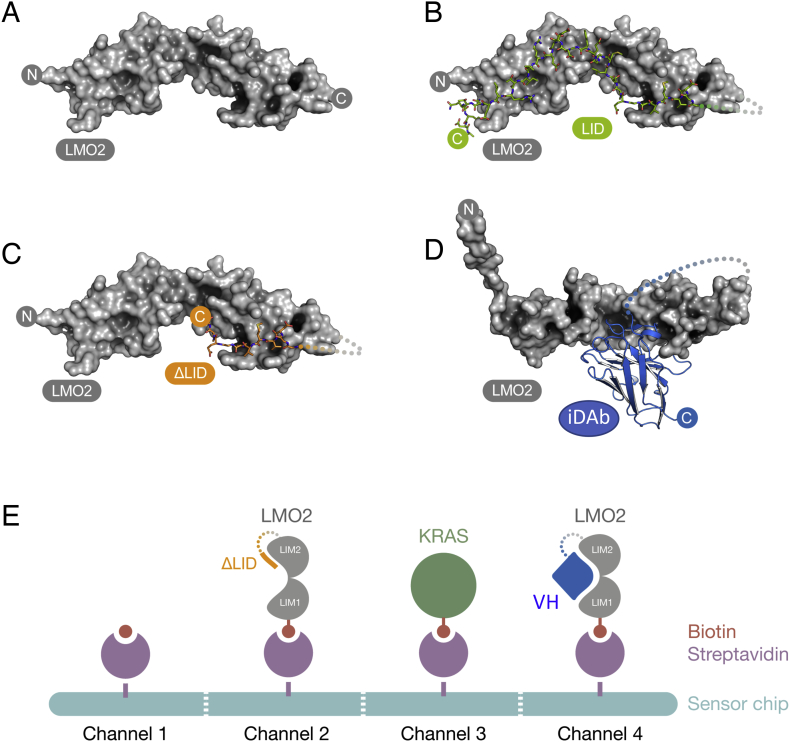
Fig. 1.
LMO2 fusion proteins and organization of cSPR chip.
Molecular models of the recombinant proteins used for the cSPR library screen. (A) LMO2 only (B) LMO2-LDB1 LID domain fusion protein (Wilkinson-White et al., 2010) (C) LMO2-ΔLID (D) LMO2-VH576 (PDB ID: 4KFZ). LMO2 is shown as a surface representation, the LDB1 LID domain as sticks and VH576 in ribbon context. The LMO2 only, LMO2-LID and LMO2-ΔLID structures are based on the LMO2-LDB1 crystal structure (PDB ID: 2XJY). The connecting glycine-serine linker was disordered in the original crystal structure and not visible, this is shown as a dotted line. The glycine-serine linker in the LMO2-VH is drawn for completion but there are no structural data. The N- and C-termini of the proteins are marked for orientation purposes.
The SPR streptavidin chips used for the cSPR screen (depicted in panel E) were channel 1: blank; channel 2: LMO2-ΔLID; channel 3: KRAS; channel 4: LMO2-gly/ser-VH.