Supplemental Digital Content is available in the text.
Keywords: acute respiratory distress syndrome, gene expression profiling, machine learning, mechanical ventilation, neutrophils, pediatric
Abstract
Objectives:
To identify differentially expressed genes and networks from the airway cells within 72 hours of intubation of children with and without pediatric acute respiratory distress syndrome. To test the use of a neutrophil transcription reporter assay to identify immunogenic responses to airway fluid from children with and without pediatric acute respiratory distress syndrome.
Design:
Prospective cohort study.
SETTING:
Thirty-six bed academic PICU.
PATIENTS:
Fifty-four immunocompetent children, 28 with pediatric acute respiratory distress syndrome, who were between 2 days to 18 years old within 72 hours of intubation for acute hypoxemic respiratory failure.
INTERVENTIONS:
None.
MEASUREMENTS AND MAIN RESULTS:
We applied machine learning methods to a Nanostring transcriptomics on primary airway cells and a neutrophil reporter assay to discover gene networks differentiating pediatric acute respiratory distress syndrome from no pediatric acute respiratory distress syndrome. An analysis of moderate or severe pediatric acute respiratory distress syndrome versus no or mild pediatric acute respiratory distress syndrome was performed. Pathway network visualization was used to map pathways from 62 genes selected by ElasticNet associated with pediatric acute respiratory distress syndrome. The Janus kinase/signal transducer and activator of transcription pathway emerged. Support vector machine performed best for the primary airway cells and the neutrophil reporter assay using a leave-one-out cross-validation with an area under the operating curve and 95% CI of 0.75 (0.63–0.87) and 0.80 (0.70–1.0), respectively.
CONCLUSIONS:
We identified gene networks important to the pediatric acute respiratory distress syndrome airway immune response using semitargeted transcriptomics from primary airway cells and a neutrophil reporter assay. These pathways will drive mechanistic investigations into pediatric acute respiratory distress syndrome. Further studies are needed to validate our findings and to test our models.
Pediatric acute respiratory distress syndrome (PARDS) is a heterogeneous syndrome with severity of lung injury defined by the degree of hypoxemia (1). Although the severity of hypoxemia is associated with outcome, it is a poor prognostic tool that does not provide insight into the complex pathobiology of PARDS (2, 3). Transcriptomics is used to explore differential gene expression patterns to reveal heterogeneity in disease samples (4, 5). Machine learning methods applied to transcriptomic data can identify important biological pathways in a disease process (6). Although gene expression of circulating leukocytes has been measured in PARDS (7–9), the airway-specific gene expression profile has not. Whether the systemic gene signature differs from the airway immune response in PARDS is unknown.
We tested a transcriptomic reporter assay using healthy donor neutrophils to assess airway fluid from a subset of intubated children to define a PARDS gene expression signature. Transcriptomic reporter assays have been used to profile the immune response of adults with sepsis, children with systemic onset juvenile idiopathic arthritis, and to establish a biomarker signature in patients prior to the onset of type 1 diabetes mellitus (10–12). Airway fluid is a valuable source of potential biomarkers (13–15) and is an attractive alternative for profiling molecular changes within the airways of intubated patients when the quantity of airway cells obtained from routine endotracheal tube suctioning and laboratory sample processing capabilities are limiting. The neutrophil transcriptomic reporter assay described provides an alternative means to profile the immune response to airway fluid from individual patients using a potentially high-throughput strategy.
The objective of this study was to explore functional gene transcript networks differentially expressed in the airway cells of intubated children with and without PARDS. We also tested a transcriptomic reporter assay using donor neutrophils treated with airway fluid to distinguish gene networks in children with and without PARDS. An analysis comparing the differential gene expression and pathways in children with moderate or severe PARDS versus no or mild PARDS was performed.
METHODS
Patient Cohort
This is an ongoing, prospective observational cohort study approved by the Emory University School of Medicine Institutional Review Board (IRB 00034236 and IRB 00113035). Informed consent was obtained prior to enrollment, and all study procedures were in accord with the with relevant guidelines and regulations in the Declaration of Helsinki. The study is being conducted at the Children’s Healthcare of Atlanta Egleston Hospital which is a 36-bed academic hospital that is affiliated with the Emory University School of Medicine. Participants in this study were enrolled between September 2018 and March 2020. Children were eligible for enrollment if they were admitted to the PICU, greater than 2 days of life, and with a corrected gestational age of 40 weeks; were less than 18 years old; and were within 72 hours of endotracheally intubation. Children were excluded if they had perinatal-related lung disease, chronic respiratory failure requiring mechanical ventilation via a tracheostomy or RAM cannula (Neotech, Valencia, CA), immunodeficiency; were receiving immunosuppression from chemotherapy for an oncologic disease; and were chronically immunosuppressed as a hematologic or solid organ transplant recipient; there was no parent or legal guardian to provide inperson written informed consent, or the attending physician did not wish the patient to participant in the study. Severity of hypoxia was classified according to the 2015 Pediatric Acute Lung Injury Consensus Conference definitions for having PARDS (16). Severity of Illness scores were determined using the Pediatric Risk of Mortality-III and Pediatric Logistic Organ Dysfunction-2 scores within 24 hours of intubation (17, 18). Duration of mechanical ventilation up to 28 days was monitored to calculate ventilator-free days.
Tracheal Aspirate Collection and Sample Processing
Tracheal aspirate samples were collected within 72 hours of endotracheal intubation with an inline Ballard suction catheter (Halyard, Alpharetta, GA) connected to a sterile Lukens trap (Covidien, Walpole, MA) using up to 5 mL of sterile saline and processed according to published protocols (19, 20). For infants and toddlers, 1–2 mL of saline is instilled. The Ballard inline suction catheter is passed once to obtain a sample. If 50% of the instilled volume is not returned, then a repeat passage of the Ballard is performed. The Ballard suction catheter (Halyard, Alpharetta, GA) may be flushed with additional saline to move the aspirate into the Lukens trap (Covidien, Walpole, MA). Cell viability was determined with trypan blue exclusion on a Countess hemocytometer. Cell purity was assessed by Shandon Kwik-Diff (Thermo Scientific, Waltham, MA) staining of cytospin cell preparations (20). Up to 1 × 106 cells were stored in RNALater at –80°C until RNA was extracted for gene expression analysis.
RNA Preparation
RNA was isolated from airway cells using the Nucleospin RNA II kit with on-column genomic DNA digestion according to the manufacturer’s protocol (Takara, Mountain View, CA). RNA sizing quantification and quality control were performed in the Emory Integrated Genomics Core on an Agilent 2100 bioanalyzer using Pico and Nano Agilent kits (Agilent Technologies, Santa Clara, CA) and a Tecan optical density plate reader (Tecan, Männedorf, Switzerland) to measure the concentration of the RNA (20). A low input RNA amplification kit was used. Due to a high signal from the B2M gene target, attenuation of this gene was performed using with synthetic oligos.
Neutrophil Isolation and Cell Culture
Blood was collected from a healthy adult donor in EDTA vacutainer tubes and centrifuged at 400× g to separate cells from platelet-rich plasma. Pelleted blood cells were resuspended in sterile phosphate buffered saline (PBS; Fisher Scientific, Waltham, MA) with 2.5 mM EDTA up to the original whole blood volume. Neutrophils were purified by negative selection from PBS-EDTA washed whole blood using the EasySep Direct Human Neutrophil Isolation kit (StemCell Technologies, Cambridge, MA) according to the manufacturer’s protocol. Neutrophils were resuspended in a 1:1 vol:vol mixture of airway supernatant (ASN) from individual patients in Roswell Park Memorial Institute 1,640 medium with L-glutamine (Corning, Corning, NY) 10% fetal calf serum supplemented with penicillin, streptomycin, and gentamicin. Neutrophils were cultured overnight in the ASN:media mixture in a humidified 37°C, 5% Co2 incubator. Neutrophils were pelleted by centrifugation at 400× g at 4°C for 15 minutes, resuspended in 500 µL of RNALater, and stored at –80 °C until RNA was isolated. A schematic of the tracheal aspirate sample processing and neutrophil transcriptomic reporter assay using airway fluid is shown in Supplementary Figure 1 (http://links.lww.com/CCX/A628).
Nanostring Array
The Human Immunology v2 NanoString nCounter Gene Expression CodeSet was used (NanoString, Seattle, WA). All NanoString-based measurements were conducted at the Emory University Integrated Genomics Core facility. The Nanostring platform is forgiving of low abundant RNA with the use of an amplification step applied to all samples and is less prone to artifact from fragmented RNA compared with traditional sequencing approaches (21).
Feature Selection and Class Prediction
Given the limited number of airway samples and high number of features, ElasticNet feature selection was implemented in Python to reduce the important features from 594 to 62 gene markers that strongly differentiated children with versus without PARDS (22). We followed our previously develop stability analysis pipeline to extract minimal redundant gene markers (23, 24). Genes with Pearson correlation coefficient greater than 0.8 were removed from the model to minimize redundancy between model features. Following recursive feature elimination and adjustment for demographic data, random forest (RF) (25), eXtreme Gradient Boosting (XGBoost) (26, 27), and support vector machines (SVMs) (28) were used to develop and evaluate binary classifiers for predicting the presence or absence of PARDS from airway cell samples and donor neutrophils treated with airway fluid from intubated patients. A flow diagram of the feature selection and modeling analysis pipeline using machine learning methods is shown in Supplementary Figure 2 (http://links.lww.com/CCX/A628).
Pathway Analysis
The R package PAthway NEtwork Visualizer (PANEV) uses Kyoto Encyclopedia of Genes and Genomes (KEGG) database to map the correlated first- and second-level pathways using the genes provided by the user (29). The genes resulted from the ElasticNet feature selection and the top 10 candidate pathways drawn from KEGG database were fed into PANEV. The PANEV program searched the KEGG database to find second-level pathways that link with the first-level pathways and the set of provided genes identified previously and visualize a comprehensive network of the genes. The same pipeline was then applied to the donor neutrophils transcriptomic reporter assay NanoString gene transcription results and was compared with the pathways identified using the NanoString gene transcription results obtained from the primary airway cells of patients.
Model Training and Validation
Models were trained using NanoString expression data on the 52 primary airway cell samples using the 62 common genes between the two separate runs of the ElasticNet algorithm. Leave-one-out cross-validation (LOOCV) was used to validate the primary airway cell model (Model 1). A second model (Model 2) was trained on the 18 donor neutrophil samples treated with airway fluid from patients with and without PARDS and validated using LOOCV. Area under the receiver operating characteristic (AUROC) curves were constructed using the scikit-learn package (30).
RESULTS
Cohort Description
Of the 52 children enrolled in the study with RNA available for analysis, there were 28 children who met PARDS criteria, and 24 children who did not meet PARDS criteria. Demographics and clinical characteristics are summarized in Table 1.
TABLE 1.
Demographic and Clinical Characteristics of Study Participants
| Characteristics | Pediatric Acute Respiratory Distress Syndrome Status | p | |
|---|---|---|---|
| No, n = 24 (46%) | Yes, n = 28 (54%) | ||
| Age (yr), median (IQR) | 0.75 (0.11–2.46) | 0.85 (0.29–2.05) | 0.34 |
| Sex, n (%) | |||
| Female/male | 11 (46)/13 (54) | 12 (43)/16 (57) | 0.83 |
| Race, n (%) | |||
| Black | 13 (54.1) | 11 (39.3) | 0.12 |
| White | 9 (37.5) | 12 (42.8) | |
| Unknown | 2 (8.4) | 1 (3.6) | |
| Multiple | 0 (0) | 4 (14.3) | |
| Ethnicity, n (%) | 0.46 | ||
| Hispanic or Latino | 2 (8.3) | 1 (3.6) | |
| Non-Hispanic or Latino | 22 (91.7) | 27 (96.4) | |
| Severity of lung injurya, n (%) | |||
| At risk | 24 (100) | NA | NA |
| Mild | NA | 10 (35.7) | |
| Moderate | NA | 10 (35.7) | |
| Severe | NA | 8 (28.6) | |
| Severity of Illness scores, median (range) | |||
| Pediatric Risk of Mortality III | 11.5 (3–23) | 14.5 (2–31) | 0.19 |
| Pediatric Logistic Organ Dysfunction | 5.5 (0–13) | 6 (3–18) | 0.14 |
| Ventilator days, median (quartile 1–quartile 3) | 3 (2–4) | 7 (3.25–11.75) | 0.0004 |
| Extracorporeal life support, n (%) | 0 (0) | 3 (10.7) | 0.0491 |
| Length of stay, median (IQR) | |||
| PICU (d) | 4 (3–7) | 9 (6–14) | 0.0002 |
| Hospital (d) | 9 (4–11.75) | 13 (8–19) | 0.01 |
| 28 d mortality, n (%) | |||
| Dead | 0 (0) | 3 (10.7) | 0.0491 |
| Viral respiratory panel, n (%) | |||
| Positive | 18 (75) | 19 (68) | 0.0032 |
| No virus detected | 0 (0) | 7 (25) | |
| Not assessed | 6 (25) | 2 (7) | |
| Respiratory culture, n (%) | |||
| No growth | 6 (25) | 3 (11) | 0.0284 |
| Bacterial growth | 13 (54) | 20 (71) | |
| Bacterial growth only | 0 (0) | 6 (21) | |
| Virus + bacterial coinfection | 13 (54) | 14 (50) | |
| Not assessed | 5 (21) | 5 (18) | |
IQR = interquartile range, NA = not applicable.
aSeverity of lung injury is defined using oxygenation index or O2 saturation index using the Pediatric Acute Lung Injury Consensus Conference definitions (16).
Comparisons were made with a Mann-Whitney U test for continuous variables or a χ2 for categorical variables.
Feature Selection, Classification, and Network Analysis of Differentially Expressed Genes From Primary Airway Cell Samples of Children With Versus Without PARDS
We applied ElasticNet to select the most important genes involved in distinguishing children with versus without PARDS. The ElasticNet model accounts for collinearity and selects for the most informative features (22). There were 62 genes selected using ElasticNet feature selection, after adjusting for demographic data, that were ranked by normalized importance coefficient in the heatmap shown in Supplementary Figure 3a (http://links.lww.com/CCX/A628). These selected genes corresponded to the top 10 KEGG pathways shown in Figure 1A and were ranked by the number of genes involved in the pathway. KEGG pathways included cytokine-cytokine receptor interactions, viral protein interactions with cytokine and cytokine receptors, chemokine signaling, autoimmune disease pathways rheumatoid arthritis, systematic lupus erythematosus, inflammatory bowel disease, and infectious processes, such as tuberculosis and Staphylococcus aureus. Although none of these children were infected with severe acute respiratory syndrome coronavirus 2, genes related to the coronavirus disease 2019 (COVID-19) pathway were discovered. The KEGG pathway labeled coronavirus/COVID-19 pathway (hsa05171) includes proinflammatory cytokines such as interleukin (IL)–6, tumor necrosis factor (TNF)–α, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB), IL-1β, IL-8, and IL-12 signaling through the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway. The COVID-19 pathway also encompasses viral pathogen recognition pathways such as Toll-like receptor signaling through MyD88 and retinoic acid-inducible gene I (RIG-1)/melanoma differentiation-associated protein 5 pathways. Furthermore, activation of the antiviral type I interferon (IFN α/β) response and the complement cascade are found within the COVID-19–labeled pathway (Supplementary Fig. 4, http://links.lww.com/CCX/A628). Additional pathways included the complement and coagulation cascades, TNF signaling, and T helper 17 (Th17) cell differentiation.
Figure 1.
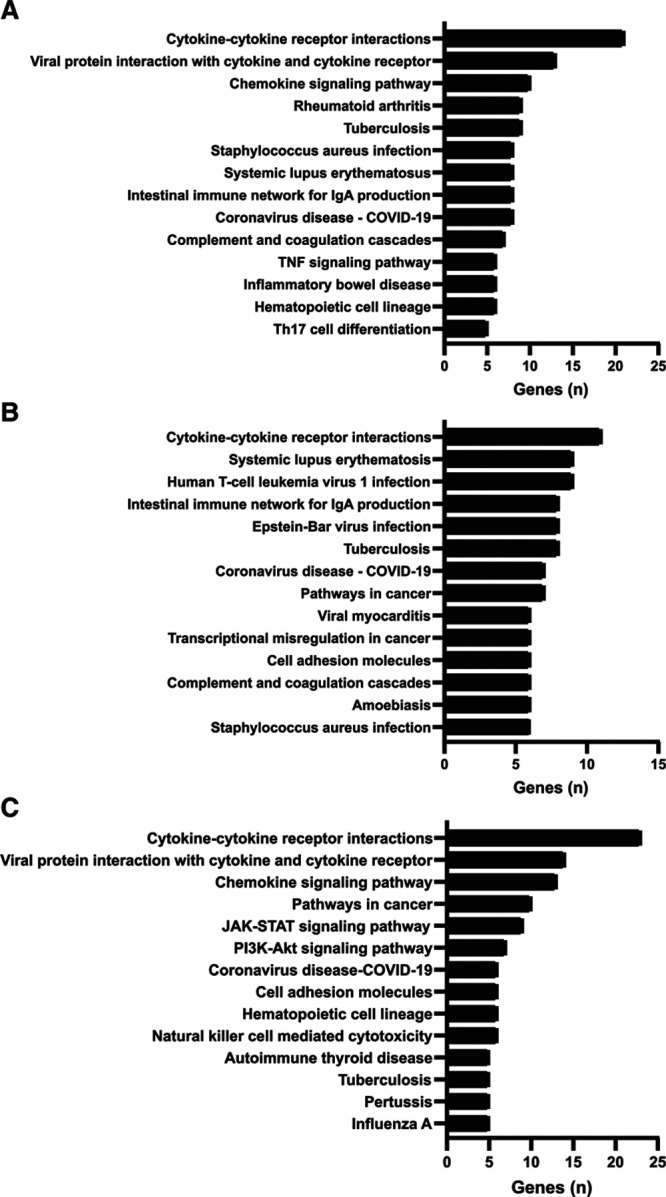
Selected genes from the top 10 KEGG pathways ranked by the number of genes involved in the pathway. A, Bar graph of the top 10 KEGG pathways identified in primary airway cells with the highest number of genes from the ElasticNet feature selection for children with versus without pediatric acute respiratory distress syndrome (PARDS). B, Bar graph of the top 10 KEGG pathways identified in primary airway cells with the highest number of genes from the ElasticNet feature selection for children with moderate/severe PARDS versus no/mild PARDS. C, Bar graph of the top 10 KEGG pathways identified in the neutrophil reporter assay with the highest number of genes from the ElasticNet feature selection for children with versus without PARDS. COVID-19 = coronavirus disease 2019, IgA = immunoglobulin A, JAK = Janus kinase, KEGG = Kyoto Encyclopedia of Genes and Genomes, PI3k-AKT = phosphoinositide 3-kinase/protein kinase B, STAT = signal transducer and activator of transcription, Th17 = T helper 17, TNF = tumor necrosis factor.
We next visualized networks of pathways involved in the primary airway cell immune response from children with versus without PARDS by imputing the list of genes selected using ElasticNet into the PANEV package in R (29). PANEV searches the KEGG library to find first-level pathways that correspond to the imputed genes discovered using machine learning methods and maps the links to second-level pathways. The top 10 first-level and second-level pathways and the corresponding genes contributing to each pathway are shown in Supplementary Figure 3b (http://links.lww.com/CCX/A628) and are listed in Supplementary File 1, (http://links.lww.com/CCX/A629) and Supplementary File 2 (http://links.lww.com/CCX/A630). In addition to the primary pathways discussed above, secondary pathways included the JAK/STAT pathway, the Toll-like receptor signaling pathway, endocytosis, apoptosis, natural killer (NK) cell mediated cytotoxicity, B-cell receptor signaling pathway, osteoclast differentiation, and antigen processing and presentation.
Feature selection, Classification, and Network Analysis o Differentially Expressed Genes From Primary Airway Cell Samples of Children With Moderate/Severe PARDS Versus No/Mild PARDS
We next performed an analysis on the differentially expressed genes from the primary airway samples comparing children with moderate or severe PARDS to those with no or mild PARDS. Differentially expressed genes corresponded to the top 10 KEGG pathways shown in Figure 1B and were ranked by the number of genes involved in the pathway. The top KEGG pathway was cytokine-cytokine receptor interaction as in the PARDS versus no PARDS analysis, and many viral infection and autoimmune pathways were the same (compare Fig. 1A with Fig. 1B). Genes were ranked by a normalized importance coefficient as shown in the heatmap in Supplementary Figure 5a (http://links.lww.com/CCX/A628). Network analysis with PANEV is shown in Supplementary Figure 5b (http://links.lww.com/CCX/A628). The top 10 genes linked to the primary and secondary pathways are listed in Supplementary File 3 (http://links.lww.com/CCX/A631) and Supplementary File 4 (http://links.lww.com/CCX/A632). The secondary network links related to endocytosis, apoptosis, cell adhesion molecules, antigen processing and presentation, Toll-like receptor signaling, JAK-STAT signaling, NK-mediated cytotoxicity, B-cell receptor signaling, and TNF signaling in children with moderate or severe PARDS versus no or mild PARDS were the same as those found in the analysis of children with versus without PARDS. Additional secondary network links in moderate or severe PARDS compared with no or mild PARDS included signaling pathways involving calcium, 3',5'-cyclic adenosine monophosphate, NF-kB, hypoxia inducible factor 1, phosphoinositide 3-kinase/protein kinase B, neutrophil extracellular trap formation, complement and coagulation cascades, platelet activation, vascular smooth muscle contraction, leukocyte transendothelial migration, and Fc gamma receptor–mediated phagocytosis.
PARDS Severity Markers Identified Using an Airway Fluid-Exposed Neutrophil Reporter Assay
We next used a neutrophil reporter assay to test whether healthy donor neutrophils would mount a similar transcriptional response when exposed to airway fluid from children with versus without PARDS. A schematic of the donor neutrophil reporter assay is illustrated in Supplementary Figure 1 (http://links.lww.com/CCX/A628). As in the primary airway cell analysis, features were selected using ElasticNet, and genes were ranked by a normalized importance coefficient as shown in the heatmap in Supplementary Figure 6a (http://links.lww.com/CCX/A628). The top 10 KEGG pathways of importance were explored and ranked by number of genes in each pathway as shown in Figure 1C. Although there were only seven genes shared between the primary airway and neutrophil reporter assay analysis, the top three pathways were the same (compare Fig. 1A with Fig. 1C). Other shared pathways included the immune response to tuberculosis, genes of the hematopoietic cell lineage, and coronavirus disease—COVID-19 networks. Network analysis with the PANEV package identified the cell adhesion and JAK/STAT pathways as primary levels in the neutrophil reporter assay that were identified as secondary levels in the primary airway cell analysis (Supplementary Figure 6b, http://links.lww.com/CCX/A628, compare with Supplementary Fig. 3b, http://links.lww.com/CCX/A628). The complement and coagulation cascades and the TNF signaling pathway that were identified as primary level pathways in the airway cells were also identified as a secondary level in the neutrophil reporter assay (Supplementary Fig. 6b, http://links.lww.com/CCX/A628, compare with Supplementary Figure 3b, http://links.lww.com/CCX/A628). Finally, the Toll-like receptor signaling and B-cell receptor signaling pathways were identified as secondary level in both the transcriptomics profiles of the airway cells and the neutrophil reporter assay experiments. Other secondary level pathways identified in the neutrophil reporter assay included the mitogen-activated protein kinase signaling pathway, calcium signaling, NF-kB signaling, extracellular membrane-receptor interactions, platelet activation, nucleotide-binding oligomerization domain-like receptor signaling, RIG-1-like receptor signaling, cytosolic DNA-sensing, T-cell receptor signaling, leukocyte transendothelial migration, and regulator of actin cytoskeleton. The top 10 first-level and second-level pathways and the corresponding genes contributing to each pathway for the neutrophil reporter assay are listed in Supplementary File 5 (http://links.lww.com/CCX/A633) and Supplementary File 6 (http://links.lww.com/CCX/A634).
Model Performance of Airway Markers Predicting PARDS
To illustrate the performance of the identified markers from primary airway cells of children with versus without PARDS, we built prediction models RF, XGBoost, and SVM algorithms (25–28). The training model characteristics predicting PARDS are shown in the top third of Table 2. A multivariable logistic regression model using the base ElasticNet selected genes showed poor performance (AUROC, 0.357; 95% CI, 0.229–0.506). Application of three machine learning algorithms improved model performance. Both the XGBoost and SVM models performed equally well with AUROC curves of 0.74 (95% CIs, 0.61–0.86) and 0.75 (95% CI, 0.63–0.87) in the primary airway cell transcriptomics assay. We then trained the models on the neutrophil reporter assay data and achieved the AUROC performance for all three models as shown in Supplementary Table 1 (http://links.lww.com/CCX/A627). The SVM model performed the best in the neutrophil donor reporter assay with an AUROC of 0.89 (95% CI, 0.7–1.0).
TABLE 2.
Test Characteristics of Pediatric Acute Respiratory Distress Syndrome Models Using Leave-One-Out Cross-Validation on Primary Airway Samples (n = 54, 28 Pediatric Acute Respiratory Distress Syndrome)
| Model | eXtreme Gradient Boosting | Random Forest | Support Vector Machine | |||
|---|---|---|---|---|---|---|
| Variables | Values | 95% CI | Values | 95% CI | Values | 95% CI |
| True negatives | 15 | — | 18 | — | 17 | — |
| False positives | 9 | — | 6 | — | 7 | — |
| True positives | 24 | — | 21 | — | 23 | — |
| False negatives | 4 | — | 7 | — | 5 | — |
| Sensitivity | 0.85 | 0.54–0.86 | 0.75 | 0.57–0.91 | 0.82 | 0.57–0.89 |
| Specificity | 0.62 | 0.53–0.93 | 0.75 | 0.50–0.87 | 0.7 | 0.54–0.91 |
| Positive predictive value | 0.72 | 0.66–0.95 | 0.77 | 0.54–0.88 | 0.76 | 0.62–0.93 |
| Negative predictive value | 0.78 | 0.40–0.80 | 0.72 | 0.53–0.83 | 0.77 | 0.49–0.86 |
| Positive likelihood ratio | 2.28 | 1.41–8.45 | 3 | 1.43–5.37 | 2.81 | 1.52–7.47 |
| Negative likelihood ratio | 0.22 | 0.19–0.61 | 0.33 | 0.14–0.64 | 0.25 | 0.15–0.59 |
| Area under the receiver operating characteristic curve | 0.74 | 0.61–0.86 | 0.7 | 0.56–0.82 | 0.75 | 0.63–0.87 |
Em dashes (—) indicate variables that do not have 95% CIs.
DISCUSSION
We used targeted immune transcriptomics and machine learning methods to discover immune pathways involved in the airway immune response of children with PARDS within 72 hours of endotracheal intubation. We identified gene networks involved in cytokine and chemokine signaling, the antiviral response, the complement and coagulation cascades, S. aureus infection, autoimmunity, and the Th17 response. We performed an analysis separating children with moderate or severe PARDS from those with no or mild PARDS and identified similar gene networks as in the PARDS versus no PARDS analysis; however, more of the genes differentially regulated were immune checkpoint pathways and wound healing/fibrosis pathways. We also report the use of a neutrophil transcription reporter assay to investigate the immunogenicity of PARDS airway fluid. We found similar and related pathway activation from primary airway cells and donor neutrophils treated with airway fluid from children with versus without PARDS. We trained and tested several machine learning models to predict PARDS. Our results are hypothesis generating and support further investigation into the pathways involved in PARDS.
Clinical and biological heterogeneity are characteristic features of PARDS. In adults machine learning algorithms applied to large datasets of well-phenotyped acute respiratory distress syndrome (ARDS) patients enrolled in randomized control trials have described two ARDS endotypes with differential responses to interventions (31–36). Whole blood transcriptomics of children enrolled for blood sampling within 24 hours of meeting PARDS criteria revealed three subphenotypes using unsupervised clustering methods (9). Despite differences in blood versus airway cell sampling, immunocompromised status, and analytical techniques, whole blood transcriptomics and airway Nanostring experiments identified common pathways such as autoimmune thyroid disease, cytokines and inflammatory response, S. aureus infection, complement and coagulation cascades, T-cell receptor signaling, and Th17 cell differentiation (9). Although endotyping was not the aim of this study, we envision endotyping of children with PARDS a future goal.
The top regulated gene in the primary airway cell transcriptomics assay is IL-17A, a key cytokine for the recruitment, activation, and migration of neutrophils to inflamed characteristic of ARDS (37). The Th17 response is characterized by a cytokine milieu that includes IL-6, transforming growth factor beta, IL-21, IL-1b, and IL-23. However, Th17 cells also have plasticity and can deviate toward a Th1 phenotype which is useful for host defense against infections and recruitment of cytotoxic lymphocytes, natural killer (NK) cells, NK T cells, and macrophages via the chemokines chemokine (C-X-C motif) ligand (CXCL)9 and CXCL10 (37, 38). Downstream signaling for Th17 differentiation depends on activation of the transcription factor STAT3 (39). The Th17 response may be amplified and dysregulated in PARDS. For example, elevated plasma and bronchoalveolar lavage fluid levels of IL-17A are found in adults with ARDS and are associated with increased percentages of neutrophils, alveolar permeability, and extrapulmonary organ dysfunction (40). An elevated ratio of Th17/regulatory T cells in adults with ARDS also predicts 28-day mortality (41). Whether these findings are also relevant to children with PARDS is unclear and warrants further study.
The transcriptomic reporter assay is grounded in the idea that metabolites, pattern-associated molecular patterns, damage-associated molecular patterns, chemokines, cytokines, and inflammatory mediators within the airway fluid will elicit an immune response in a genetically homogeneous pool of donor neutrophils. Neutrophils were chosen because they are the predominant innate immune cell recruited and recovered from tracheal aspirate lavage of the airways of children with PARDS (19, 20, 42, 43). Neutrophils have the greatest differential gene expression response compared with monocytes and monocyte-derived dendritic cells in response to the plasma of septic adults (10). Although few genes were shared between the primary airway cell and neutrophil reporter assay analysis, there was overlap in the pathways that emerged.
The top regulated gene was early growth response (Egr)–1, an immediate-early response transcription factor with a key role in inflammation, extracellular matrix formation, thrombosis, apoptosis, and fibrosis, that are involved in acute lung injury (44–46). For example, suppression of Egr-1 following immune complex-induced acute lung inflammation in mice with the peroxisome proliferator-activated receptor-γ agonist rosiglitazone reduced proinflammatory cytokine expression (47). Inhibition of the mitogen-activated protein kinase kinase/extracellular signal-regulated kinase/early growth response protein 1 pathway decreased lung edema, inflammation, and neutrophil migration in a lipopolysaccharide-induced mouse model of acute lung injury (48). There are several other notable genes in the moderate/severe PARDS groups that are implicated in acute lung injury including CTLA4, ICOS, CD27 which are important in immune checkpoint molecules that regulate T-cell behavior (49, 50), Abl-1 a tyrosine kinase which regulates vascular leak in sepsis-triggered ARDS, and arginase 1 an enzyme that metabolizes arginine and may contribute to pulmonary capillary vascular leak, T-cell modulation by neutrophils secreting arginase 1, and collagen deposition leading to fibrosis (51–56).
The top up-regulated gene identified in the neutrophil reporter assay is NT5E, also known as cluster of differentiation 73 (CD73), which is an enzyme that converts danger signals such as adenosine triphosphate, adenosine diphosphate, and adenosine monophosphate to adenosine (57). Adenosine is an anti-inflammatory metabolite that regulates immune tolerance through immunosuppressive regulatory T cells and anergic CD4 T cells (58). Adenosine limits the inflammatory response of neutrophils through a negative feedback mechanism with adenosine receptors expressed on neutrophils (59). In preclinical models of ARDS, up-regulation of CD73 on epithelial and endothelial cells by IFN β-1a can prevent vascular leakage and inhibit leukocyte recruitment (60–64). Unfortunately, a recent phase III randomized control trial of IFN β-1a showed no difference in a composite outcome of death or ventilator-free days over 28 days in adults with moderate to severe ARDS (57). Nevertheless, the finding of CD73 in the neutrophil reporter assay of PARDS samples supports the relevance of this finding in our study.
Machine learning algorithms are becoming more routinely used to analyze differential gene expression in het-erogeneous conditions such as sepsis and ARDS (31, 32). Due to high dimensionality of the dataset, we applied the ElasticNet feature selection method to select robust features, while minimizing collinearity. This resulted in a reduction to a set of 49 important genes that differentiated the two cohorts. Among our classifiers, XGBoosst and SVM were equally robust in predicting PARDS using airway samples, with the SVM model particularly outperforming when applied to neutrophil donor dataset. XGBoost, as a scalable gradient tree boosting algorithm (26), gradually creates a model by optimizing a random differentiable loss function (27). SVM enhances the geometric characteristics of the training data to find the exact hyperplane that separates the two classes (28). Several studies have demonstrated the robustness of SVM models to outperform more sophisticated training algorithms when evaluated again high-dimensional data (65), consistent with our observations in this study.
Our study is limited by enrollment of children from a single-center, which may limit generalizability. Our limited sample size did not enable us to model for important clinical outcomes such as new functional disability, ventilator-free days, or mortality. We only collected a single sample within a 72-hour collection window following intubation using nonstandardized collection volumes in the clinical course and therefore cannot correlate clinical trajectory with changes in airway immune response. By excluding known immunocompromised children from our study, we cannot comment on how an immunocompromised state effects the airway immune response in PARDS. Despite our limited sample size, we attempted to determine differences in gene expression among children with no detected infectious trigger without PARDS and those with a viral or any infectious trigger (bacterial, viral, or both) with PARDS, but we did not find any differential genes selected using ElasticNet. Validation of our results with an external cohort is needed to assess reproducibility of the PARDS airway immune response reported. Larger well-phenotyped pediatric cohorts are needed to identify subclasses of children who may differentially respond to targeted therapies and interventions.
In conclusion, we report an airway immune response gene expression signature in intubated children with PARDS using a Nanostring platform and machine learning algorithms. Nanostring airway immune response profiling may provide a clinically useful strategy to understand the complex heterogeneity of the immune response of PARDS and to endotype patients for prognostic and predictive enrichment in PARDS trials.
ACKNOWLEDGMENTS
We thank the bedside caregivers of the patients involved in this study for their skilled and compassionate care.
Supplementary Material
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
Drs. Grunwell, Fitzpatrick, and Kamaleswaran conceived and developed the study, supervised the acquisition of the biological data, and analyzed and interpreted the data. Dr. Grunwell drafted and edited the article. Drs. Rad, Fitzpatrick, and Kamaleswaran assisted with drafting and editing the article. Drs. Stephenson and Mohammad helped with patient sample processing, performed experiments, and edited the article. Dr. Opolka assisted in identifying, consenting, acquiring patient samples and assisted in collecting clinical information about the patients. All authors edited and approved the final version of this article.
Supported, in part, by the National Institutes of Health (NIH) grants K12HD072245 (Atlanta Pediatric Scholars Program), K23 HL151897-01 and an Emory University Pediatrics Research Alliance Junior Faculty Focused Pilot award (to Dr. Grunwell). Also supported, in part, by the NIH grant K24 NR018866 (to Dr. Fitzpatrick). Supported, in part, by the Emory Integrated Genomics Core, which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the Georgia Clinical and Translational Science Alliance of the National Institutes of Health under Award Number UL1TR002378.
The authors have disclosed that they do not have any potential conflicts of interest.
The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
This work was performed at Emory University School of Medicine, Children’s Healthcare of Atlanta, and Georgia Institute of Technology.
REFERENCES
- 1.Khemani RG, Smith L, Lopez-Fernandez YM, et al. ; Pediatric Acute Respiratory Distress syndrome Incidence and Epidemiology (PARDIE) Investigators; Pediatric Acute Lung Injury and Sepsis Investigators (PALISI) Network. Paediatric acute respiratory distress syndrome incidence and epidemiology (PARDIE): An international, observational study. Lancet Respir Med. 2019; 7:115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ranieri VM, Rubenfeld GD, Thompson BT, et al. ; ARDS Definition Task Force. Acute respiratory distress syndrome: The Berlin definition. JAMA. 2012; 307:2526–2533 [DOI] [PubMed] [Google Scholar]
- 3.Zinter MS, Orwoll BE, Spicer AC, et al. Incorporating inflammation into mortality risk in pediatric acute respiratory distress syndrome. Crit Care Med. 2017; 45:858–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer NJ, Calfee CS. Novel translational approaches to the search for precision therapies for acute respiratory distress syndrome. Lancet Respir Med. 2017; 5:512–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bos LDJ, Scicluna BP, Ong DSY, et al. Understanding heterogeneity in biologic phenotypes of acute respiratory distress syndrome by leukocyte expression profiles. Am J Respir Crit Care Med. 2019; 200:42–50 [DOI] [PubMed] [Google Scholar]
- 6.Abbas M, El-Manzalawy Y. Machine learning based refined differential gene expression analysis of pediatric sepsis. BMC Med Genomics. 2020; 13:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yehya N, Thomas NJ, Wong HR. Evidence of endotypes in pediatric acute hypoxemic respiratory failure caused by sepsis. Pediatr Crit Care Med. 2019; 20:110–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yehya N, Wong HR. Adaptation of a biomarker-based sepsis mortality risk stratification tool for pediatric acute respiratory distress syndrome. Crit Care Med. 2018; 46:e9–e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yehya N, Varisco BM, Thomas NJ, et al. Peripheral blood transcriptomic sub-phenotypes of pediatric acute respiratory distress syndrome. Crit Care. 2020; 24:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khaenam P, Rinchai D, Altman MC, et al. A transcriptomic reporter assay employing neutrophils to measure immunogenic activity of septic patients’ plasma. J Transl Med. 2014; 12:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Jia S, Geoffrey R, et al. Identification of a molecular signature in human type 1 diabetes mellitus using serum and functional genomics. J Immunol. 2008; 180:1929–1937 [DOI] [PubMed] [Google Scholar]
- 12.Pascual V, Allantaz F, Arce E, et al. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J Exp Med. 2005; 201:1479–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers AJ, Contrepois K, Wu M, et al. Profiling of ARDS pulmonary edema fluid identifies a metabolically distinct subset. Am J Physiol Lung Cell Mol Physiol. 2017; 312:L703–L709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aggarwal A, Baker CS, Evans TW, et al. G-CSF and IL-8 but not GM-CSF correlate with severity of pulmonary neutrophilia in acute respiratory distress syndrome. Eur Respir J. 2000; 15:895–901 [DOI] [PubMed] [Google Scholar]
- 15.Grégoire M, Uhel F, Lesouhaitier M, et al. Impaired efferocytosis and neutrophil extracellular trap clearance by macrophages in ARDS. Eur Respir J. 2018; 52:1702590. [DOI] [PubMed] [Google Scholar]
- 16.Khemani RG, Smith LS, Zimmerman JJ, et al. ; Pediatric Acute Lung Injury Consensus Conference Group. Pediatric acute respiratory distress syndrome: definition, incidence, and epidemiology: Proceedings from the pediatric acute lung injury consensus conference. Pediatr Crit Care Med. 2015; 16:S23–S40 [DOI] [PubMed] [Google Scholar]
- 17.Leteurtre S, Duhamel A, Salleron J, et al. ; Groupe Francophone de Réanimation et d’Urgences Pédiatriques (GFRUP). PELOD-2: An update of the pediatric logistic organ dysfunction score. Crit Care Med. 2013; 41:1761–1773 [DOI] [PubMed] [Google Scholar]
- 18.Pollack MM, Patel KM, Ruttimann UE. PRISM III: An updated pediatric risk of mortality score. Crit Care Med. 1996; 24:743–752 [DOI] [PubMed] [Google Scholar]
- 19.Grunwell JR, Giacalone VD, Stephenson S, et al. Neutrophil dysfunction in the airways of children with acute respiratory failure due to lower respiratory tract viral and bacterial coinfections. Sci Rep. 2019; 9:2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunwell JR, Stephenson ST, Mohammad AF, et al. Differential type I interferon response and primary airway neutrophil extracellular trap release in children with acute respiratory distress syndrome. Sci Rep. 2020; 10:19049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veldman-Jones MH, Brant R, Rooney C, et al. Evaluating robustness and sensitivity of the NanoString Technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res. 2015; 75:2587–2593 [DOI] [PubMed] [Google Scholar]
- 22.Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Ser B Stat Methodol. 2005; 67:301–320 [Google Scholar]
- 23.Mohammed A, Cui Y, Mas VR, et al. Differential gene expression analysis reveals novel genes and pathways in pediatric septic shock patients. Sci Rep. 2019; 9:11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee S, Mohammed A, Wong HR, et al. Machine learning identifies complicated sepsis course and subsequent mortality based on 20 genes in peripheral blood immune cells at 24 H post-ICU admission. Front Immunol. 2021; 12:592303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svetnik V, Liaw A, Tong C, et al. Random forest: A classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003; 43:1947–1958 [DOI] [PubMed] [Google Scholar]
- 26.Chen T, Guestrin C. XGBoost. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data MiningACM, San Francisco, CA, August 13–18, 2016 [Google Scholar]
- 27.Batra A, Jawa V. Classification of arrhythmia using conjunction of machine learning algorithms and ECG diagnostic criteria. Int J Biol Biomed. 2016; 1:1–7 [Google Scholar]
- 28.Stitson MO, Weston JAE, Gammerman A, et al. Theory of support vector machines. Technical Report CSD-TR-96-17. Egham Surrey, England, Royal Holloway, University of London, 1996; 117.827 188–191 [Google Scholar]
- 29.Palombo V, Milanesi M, Sferra G, et al. PANEV: An R package for a pathway-based network visualization. BMC Bioinformatics. 2020; 21:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011; 12:2825–2830 [Google Scholar]
- 31.Sinha P, Churpek MM, Calfee CS. Machine learning classifier models can identify acute respiratory distress syndrome phenotypes using readily available clinical data. Am J Respir Crit Care Med. 2020; 202:996–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinha P, Delucchi KL, McAuley DF, et al. Development and validation of parsimonious algorithms to classify acute respiratory distress syndrome phenotypes: A secondary analysis of randomised controlled trials. Lancet Respir Med. 2020; 8:247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calfee CS, Delucchi K, Parsons PE, et al. ; NHLBI ARDS Network. Subphenotypes in acute respiratory distress syndrome: Latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014; 2:611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bos LD, Schouten LR, van Vught LA, et al. ; MARS consortium. Identification and validation of distinct biological phenotypes in patients with acute respiratory distress syndrome by cluster analysis. Thorax. 2017; 72:876–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha P, Delucchi KL, Thompson BT, et al. ; NHLBI ARDS Network. Latent class analysis of ARDS subphenotypes: A secondary analysis of the statins for acutely injured lungs from sepsis (SAILS) study. Intensive Care Med. 2018; 44:1859–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Famous KR, Delucchi K, Ware LB, et al. ; ARDS Network. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med. 2017; 195:331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Annunziato F, Romagnani C, Romagnani S. The 3 major types of innate and adaptive cell-mediated effector immunity. J Allergy Clin Immunol. 2015; 135:626–635 [DOI] [PubMed] [Google Scholar]
- 38.Tokunaga R, Zhang W, Naseem M, et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - a target for novel cancer therapy. Cancer Treat Rev. 2018; 63:40–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stockinger B, Veldhoen M, Martin B. Th17 T cells: Linking innate and adaptive immunity. Semin Immunol. 2007; 19:353–361 [DOI] [PubMed] [Google Scholar]
- 40.Mikacenic C, Hansen EE, Radella F, et al. Interleukin-17A Is associated with alveolar inflammation and poor outcomes in acute respiratory distress syndrome. Crit Care Med. 2016; 44:496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu ZX, Ji MS, Yan J, et al. The ratio of Th17/Treg cells as a risk indicator in early acute respiratory distress syndrome. Crit Care. 2015; 19:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grommes J, Soehnlein O. Contribution of neutrophils to acute lung injury. Mol Med. 2011; 17:293–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiland JE, Davis WB, Holter JF, et al. Lung neutrophils in the adult respiratory distress syndrome. Clinical and pathophysiologic significance. Am Rev Respir Dis. 1986; 133:218–225 [DOI] [PubMed] [Google Scholar]
- 44.Bhattacharyya S, Wu M, Fang F, et al. Early growth response transcription factors: Key mediators of fibrosis and novel targets for anti-fibrotic therapy. Matrix Biol. 2011; 30:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ngiam N, Post M, Kavanagh BP. Early growth response factor-1 in acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2007; 293:L1089–L1091 [DOI] [PubMed] [Google Scholar]
- 46.Dong WW, Feng Z, Zhang YQ, et al. Potential mechanism and key genes involved in mechanical ventilation and lipopolysaccharide-induced acute lung injury. Mol Med Rep. 2020; 22:4265–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan C, Chen J, Ding Y, et al. The crucial role of PPARγ-Egr-1-Pro-inflammatory mediators axis in IgG immune complex-induced acute lung injury. Front Immunol. 2021; 12:634889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen S, Xu H, Ye P, et al. Trametinib alleviates lipopolysaccharide-induced acute lung injury by inhibiting the MEK-ERK-Egr-1 pathway. Int Immunopharmacol. 2020; 80:106152. [DOI] [PubMed] [Google Scholar]
- 49.Nakajima T, Suarez CJ, Lin KW, et al. T cell pathways involving CTLA4 contribute to a model of acute lung injury. J Immunol. 2010; 184:5835–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y, Tang L, Lin M, et al. Expression of cytotoxic T-lymphocyte antigen 4 on CD4+ and CD8+ T cells is increased in acute lung injury. DNA Cell Biol. 2013; 32:722–726 [DOI] [PubMed] [Google Scholar]
- 51.Lucas R, Czikora I, Sridhar S, et al. Arginase 1: An unexpected mediator of pulmonary capillary barrier dysfunction in models of acute lung injury. Front Immunol. 2013; 4:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacobsen LC, Theilgaard-Mönch K, Christensen EI, et al. Arginase 1 is expressed in myelocytes/metamyelocytes and localized in gelatinase granules of human neutrophils. Blood. 2007; 109:3084–3087 [DOI] [PubMed] [Google Scholar]
- 53.Rotondo R, Bertolotto M, Barisione G, et al. Exocytosis of azurophil and arginase 1-containing granules by activated polymorphonuclear neutrophils is required to inhibit T lymphocyte proliferation. J Leukoc Biol. 2011; 89:721–727 [DOI] [PubMed] [Google Scholar]
- 54.Uhel F, Azzaoui I, Grégoire M, et al. Early expansion of circulating granulocytic myeloid-derived suppressor cells predicts development of nosocomial infections in patients with sepsis. Am J Respir Crit Care Med. 2017; 196:315–327 [DOI] [PubMed] [Google Scholar]
- 55.Reizine F, Lesouhaitier M, Gregoire M, et al. SARS-CoV-2-induced ARDS associates with MDSC expansion, lymphocyte dysfunction, and arginine shortage. J Clin Immunol. 2021; 41:515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maarsingh H, Pera T, Meurs H. Arginase and pulmonary diseases. Naunyn Schmiedebergs Arch Pharmacol. 2008; 378:171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ranieri VM, Pettila V, Karvonen MK, et al. Effect of intravenous interferon beta-1a on death and days free from mechanical ventilation among patients with moderate to severe acute respiratory distress syndrome: A randomized clinical trial. JAMA. 2020; 323:725–733 [DOI] [PubMed] [Google Scholar]
- 58.Bynoe MS, Viret C. Foxp3+CD4+ T cell-mediated immunosuppression involves extracellular nucleotide catabolism. Trends Immunol. 2008; 29:99–102 [DOI] [PubMed] [Google Scholar]
- 59.Kordaß T, Osen W, Eichmüller SB. Controlling the immune suppressor: Transcription factors and microRNAs regulating CD73/NT5E. Front Immunol. 2018; 9:813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eckle T, Füllbier L, Wehrmann M, et al. Identification of ectonucleotidases CD39 and CD73 in innate protection during acute lung injury. J Immunol. 2007; 178:8127–8137 [DOI] [PubMed] [Google Scholar]
- 61.Grünewald JK, Ridley AJ. CD73 represses pro-inflammatory responses in human endothelial cells. J Inflamm (Lond). 2010; 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiss J, Yegutkin GG, Koskinen K, et al. IFN-beta protects from vascular leakage via up-regulation of CD73. Eur J Immunol. 2007; 37:3334–3338 [DOI] [PubMed] [Google Scholar]
- 63.Eckle T, Faigle M, Grenz A, et al. A2B adenosine receptor dampens hypoxia-induced vascular leak. Blood. 2008; 111:2024–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eckle T, Grenz A, Laucher S, et al. A2B adenosine receptor signaling attenuates acute lung injury by enhancing alveolar fluid clearance in mice. J Clin Invest. 2008; 118:3301–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang S, Cai N, Pacheco PP, et al. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics. 2018; 15:41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


